For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTSLRRVAVAIMVLIVLLLANATVTQVFAADGLRADPRNQRVLLDEYSRQRGQITAGGQLLAYSVSTDGR FRYLRVYPNPQAYAPVTGFYSLGYSSTGLERAEDAVLNGSDERLFGRRLADFFTGRDPRGGNVDTTIKPQ VQQAAWDAMQNGCDGPCRGSVVALEPSTGKILAMVSAPSYDPNLLATHDLAAQADAWEKLRDDPQSPLLN RAISETYPPGSTFKVITTAAALQAGARPQTQLTSAPRTPLPDSTATLENFGGAPCGPGPTVSLQEAFAKS CNTAFVELGLSTGTDKLKAMAQAFGLDTPPPAIPLQVAESTTGPIVDAAALGMSSIGQRDVALTPLQNAQ VAATIANDGIAMRPYLVESLKGPDLATISTTTPEQERRAVSPQVAATLTDLMVAAEQVTQQKGAIAGVQI ASKTGTAEHGTDPRNTPPHAWYIAFAPAQDPKVAVAVLVEDGGDRLSATGGALAAPIGRATIAAALREGS *
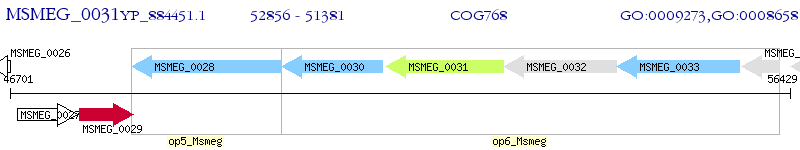
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0031 | - | - | 100% (491) | penicillin binding protein transpeptidase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4233 | - | 2e-26 | 26.98% (430) | penicillin binding protein transpeptidase domain-containing |
| M. smegmatis MC2 155 | MSMEG_6319 | - | 2e-18 | 30.73% (358) | penicillin-binding protein, transpeptidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0016c | pbpA | 0.0 | 80.33% (488) | penicillin-binding protein PbpA |
| M. gilvum PYR-GCK | Mflv_0810 | - | 0.0 | 82.48% (491) | penicillin-binding protein, transpeptidase |
| M. tuberculosis H37Rv | Rv0016c | pbpA | 0.0 | 80.33% (488) | penicillin-binding protein PbpA |
| M. leprae Br4923 | MLBr_00018 | pbpA | 0.0 | 79.47% (492) | putative penicillin-binding protein |
| M. abscessus ATCC 19977 | MAB_0035c | - | 0.0 | 69.44% (504) | penicillin-binding protein PbpA |
| M. marinum M | MMAR_0018 | pbpA | 0.0 | 83.40% (488) | penicillin-binding protein PbpA |
| M. avium 104 | MAV_0020 | - | 0.0 | 80.04% (491) | PbpA protein |
| M. thermoresistible (build 8) | TH_0586 | - | 1e-20 | 26.26% (438) | Probable penicillin-binding membrane protein pbpB |
| M. ulcerans Agy99 | MUL_0020 | pbpA | 0.0 | 82.58% (488) | penicillin-binding protein PbpA |
| M. vanbaalenii PYR-1 | Mvan_0025 | - | 0.0 | 83.10% (491) | penicillin-binding protein, transpeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0810|M.gilvum_PYR-GCK MNTSLRRIS-----------------------------------VMIMAL
Mvan_0025|M.vanbaalenii_PYR-1 MNTSLRRIS-----------------------------------VMIMAL
MSMEG_0031|M.smegmatis_MC2_155 MNTSLRRVA-----------------------------------VAIMVL
Mb0016c|M.bovis_AF2122/97 MNASLRRIS-----------------------------------VTVMAL
Rv0016c|M.tuberculosis_H37Rv MNASLRRIS-----------------------------------VTVMAL
MMAR_0018|M.marinum_M MNASLRRIS-----------------------------------VTVMAL
MUL_0020|M.ulcerans_Agy99 MNASLRRIS-----------------------------------VTVMAL
MAV_0020|M.avium_104 MNASLRRIS-----------------------------------VTVMAL
MLBr_00018|M.leprae_Br4923 MNTSLRRIS-----------------------------------VTVMAL
MAB_0035c|M.abscessus_ATCC_199 MNTSIRRVS-----------------------------------LAVMGL
TH_0586|M.thermoresistible__bu MSRAKRQAGQRGEGRSARARRTRQIVTETGAQTASFVFRHRIGNAVIFLV
*. : *: . :: :
Mflv_0810|M.gilvum_PYR-GCK IVVLLANATLTQVFTADGLRSDPRNQRVLLDEYSRQRGQISAG-------
Mvan_0025|M.vanbaalenii_PYR-1 VVVLLANATLTQVFTADGLRSDPRNQRVLLDEYSRQRGQISAG-------
MSMEG_0031|M.smegmatis_MC2_155 IVLLLANATVTQVFAADGLRADPRNQRVLLDEYSRQRGQITAG-------
Mb0016c|M.bovis_AF2122/97 IVLLLLNATMTQVFTADGLRADPRNQRVLLDEYSRQRGQITAG-------
Rv0016c|M.tuberculosis_H37Rv IVLLLLNATMTQVFTADGLRADPRNQRVLLDEYSRQRGQITAG-------
MMAR_0018|M.marinum_M IVLLLLNATMTQVFTADGLRADPRNQRVLLDEYSRQRGQITAG-------
MUL_0020|M.ulcerans_Agy99 IVLLLLNATMTPVFTADGLRADPRNQRVLLDEYSRQRGQITAG-------
MAV_0020|M.avium_104 IVLLLLNATMTQVFAADSLRADPRNQRVLLDEYSRQRGQIVAG-------
MLBr_00018|M.leprae_Br4923 IVLLLLNATVTQVFTADGLRADPRNQRVLLDEYSRQRGQITAS-------
MAB_0035c|M.abscessus_ATCC_199 VVLLLANATITQVFTAPGLRADPRNQRILLDEYSRQRGQISAS-------
TH_0586|M.thermoresistible__bu LVVVAAQLFNLQVPRAPALRAEAAGQLKVTDVEPAMRGAIVDRDFDKLAF
:*:: : * * .**::. .* : * . ** *
Mflv_0810|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0025|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0031|M.smegmatis_MC2_155 --------------------------------------------------
Mb0016c|M.bovis_AF2122/97 --------------------------------------------------
Rv0016c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0018|M.marinum_M --------------------------------------------------
MUL_0020|M.ulcerans_Agy99 --------------------------------------------------
MAV_0020|M.avium_104 --------------------------------------------------
MLBr_00018|M.leprae_Br4923 --------------------------------------------------
MAB_0035c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0586|M.thermoresistible__bu TIESRALTFQPQRIRKQLVEAKEKKSDAPEPADRLTEIAHGVAEKLNHKP
Mflv_0810|M.gilvum_PYR-GCK -GQLLAYSVSTEGRFRFLRVYPNPMAYAPVTGFYSLTYSSS---------
Mvan_0025|M.vanbaalenii_PYR-1 -GQLLAYSVSTQGRFRFLRVYPDPMIYAPVTGFYSLSYSST---------
MSMEG_0031|M.smegmatis_MC2_155 -GQLLAYSVSTDGRFRYLRVYPNPQAYAPVTGFYSLGYSST---------
Mb0016c|M.bovis_AF2122/97 -GQLLAYSVATDGRFRFLRVYPNPEVYAPVTGFYSLRYSST---------
Rv0016c|M.tuberculosis_H37Rv -GQLLAYSVATDGRFRFLRVYPNPEVYAPVTGFYSLRYSST---------
MMAR_0018|M.marinum_M -GQLLAYSVATDSRFRFLRVYPNPEVYAPVTGFYSLRYSST---------
MUL_0020|M.ulcerans_Agy99 -GQLLAYSVATDGRFRFLRVYPNPEVYAPVTGFYSLRYSST---------
MAV_0020|M.avium_104 -GQLLAYSVATDNRFRFLRVYPNPAQYAPVTGFYSLRYSST---------
MLBr_00018|M.leprae_Br4923 -GQLLAYSVATDNRFRFLRVYPNPAVYAPITGFYSLRYSST---------
MAB_0035c|M.abscessus_ATCC_199 -GQLLASSVATDSRFRFLRQYADPEVYAPVTGFYSLNYSST---------
TH_0586|M.thermoresistible__bu DATSLLKKLMSNETFVYLARAVDPAVASEITREFPEVGSERQDLRQYPGG
. * .: :: * :* :* : :* :. *.
Mflv_0810|M.gilvum_PYR-GCK ------------------GLERAEDTILNGSDERLFGRRLADFFTGRDPR
Mvan_0025|M.vanbaalenii_PYR-1 ------------------GLERAEDTILNGSDERLFGRRLADFFTGRDPR
MSMEG_0031|M.smegmatis_MC2_155 ------------------GLERAEDAVLNGSDERLFGRRLADFFTGRDPR
Mb0016c|M.bovis_AF2122/97 ------------------ALERAEDPILNGSDRRLFGRRLADFFTGRDPR
Rv0016c|M.tuberculosis_H37Rv ------------------ALERAEDPILNGSDRRLFGRRLADFFTGRDPR
MMAR_0018|M.marinum_M ------------------GLERAEDPLLNGSDQRLFGRRLADFFTGRDPR
MUL_0020|M.ulcerans_Agy99 ------------------GLERAEDPLLNGSDQRLFGRRLADFFTGRDPR
MAV_0020|M.avium_104 ------------------GLERAEDPLLNGSDERLFGRRLADFFTGRDPR
MLBr_00018|M.leprae_Br4923 ------------------GLEQAEDALLNGSDERLFGRRLADFFTGRDPR
MAB_0035c|M.abscessus_ATCC_199 ------------------GLERAEDTMLNGSDPRLFGRRLADFFTGRDPR
TH_0586|M.thermoresistible__bu SLAANIIGGIDWDGHGLLGLEDALDAVLAGTDGSITYDRGSDGVVIPGSY
.** * *.:* *:* : * :* .. ..
Mflv_0810|M.gilvum_PYR-GCK GGNVSTTINPDVQQAAWDAMEDGCNG-PCKGSVVALEPSTGKILAMVSAP
Mvan_0025|M.vanbaalenii_PYR-1 GGTVATTINPQVQRAAWDAMEKGCDG-PCTGSVVALEPSTGKILALVSAP
MSMEG_0031|M.smegmatis_MC2_155 GGNVDTTIKPQVQQAAWDAMQNGCDG-PCRGSVVALEPSTGKILAMVSAP
Mb0016c|M.bovis_AF2122/97 GGNVDTTINPRIQQAGWDAMQQGCYG-PCKGAVVALEPSTGKILALVSSP
Rv0016c|M.tuberculosis_H37Rv GGNVDTTINPRIQQAGWDAMQQGCYG-PCKGAVVALEPSTGKILALVSSP
MMAR_0018|M.marinum_M GGNVDTTINPRIQQAGWDAMQQGCGG-PCKGAVVALEPSTGKILALVSSP
MUL_0020|M.ulcerans_Agy99 GGNVDTAINPRIQQAGWDAMQQGCGG-PCKGAVVALEPSTGKILALVSSP
MAV_0020|M.avium_104 GANVDTTIRPRVQQAAWDGMQQGCGGPPCKGAVVALEPSTGKILAMVSSP
MLBr_00018|M.leprae_Br4923 GGNVDTTINPRVQQTGWDAMQQGCGGSPCKGAVVALEPSTGKILAMVSTP
MAB_0035c|M.abscessus_ATCC_199 GGSVVTTIRPDVQQAAYDAMHRGCKD-GCRGAVVALEPSTGKILAMVSTP
TH_0586|M.thermoresistible__bu RNRHEAVDGSTVQLTIDDDIQFHVQQ----QVQLAKDASGAKNVSAVVLD
:. . :* : * :. :* :.* .* :: *
Mflv_0810|M.gilvum_PYR-GCK SYDPNLLASHNIDEQAQTWQALRDNPDSPLLNRAISETYPPGSTFKVLTT
Mvan_0025|M.vanbaalenii_PYR-1 SYDPNLLATHDVAEQAEAWQRLRDDPGSPLLNRAISEAYPPGSTFKVITT
MSMEG_0031|M.smegmatis_MC2_155 SYDPNLLATHDLAAQADAWEKLRDDPQSPLLNRAISETYPPGSTFKVITT
Mb0016c|M.bovis_AF2122/97 SYDPNLLASHNPEVQAQAWQRLGDNPASPLTNRAISETYPPGSTFKVITT
Rv0016c|M.tuberculosis_H37Rv SYDPNLLASHNPEVQAQAWQRLGDNPASPLTNRAISETYPPGSTFKVITT
MMAR_0018|M.marinum_M SYDPNLLASHDPEAQAEAWKDLRDDPSSPLTNRAISETYPPGSTFKVITT
MUL_0020|M.ulcerans_Agy99 SYDPNLLASHDPEAQAEAWKDLRDDPSSALTNRAISETYPPGSTFKVITT
MAV_0020|M.avium_104 SYDPNLLSSHDPEVQAQAWQRLRDDPDNPMTNRAISETYPPGSTFKVITT
MLBr_00018|M.leprae_Br4923 SYDPNLLASHNPEEQAQAWRRLHDDPNSPLINRAISETYPPGSTFKVITT
MAB_0035c|M.abscessus_ATCC_199 SYDPNLLASQDPEKENAAWKRLTEDPAQPMLNRAIAQTYPPGSTFKVITT
TH_0586|M.thermoresistible__bu ARTAEVLAMAN-DNTFDPSQDIGRQGDRQMGNLPVTSPFEPGSVQKLVTA
: .::*: : . . : : : * .::..: ***. *::*:
Mflv_0810|M.gilvum_PYR-GCK AAALEAGATPQTQLTAAPQIPLPNSTATLENFGGTACGGGPTATLEYAFA
Mvan_0025|M.vanbaalenii_PYR-1 AAALQAGATPDTQLTAAPQIALPDSTATLQNFGGVSCGAGPTTSLRNAFA
MSMEG_0031|M.smegmatis_MC2_155 AAALQAGARPQTQLTSAPRTPLPDSTATLENFGGAPCGPGPTVSLQEAFA
Mb0016c|M.bovis_AF2122/97 AAALAAGATETEQLTAAPTIPLPGSTAQLENYGGAPCGDEPTVSLREAFV
Rv0016c|M.tuberculosis_H37Rv AAALAAGATETEQLTAAPTIPLPGSTAQLENYGGAPCGDEPTVSLREAFV
MMAR_0018|M.marinum_M AAALQAGATENDQLTAAANIPLPDSTATLENYGGAPCDSGQTVSLREAFA
MUL_0020|M.ulcerans_Agy99 AAALQAGATENDQLTAAANIPLPDSTATLENYGGAPCGSGQTVSLREAFA
MAV_0020|M.avium_104 AAALQAGASDTEQLTAAPSIPLPNSTATLENYGGQACGNDPTVSLQQAFA
MLBr_00018|M.leprae_Br4923 TAALQAGATTSDQLTAEPTIPLPGSTATLENYGGVSCGPGPTVSLSQAFA
MAB_0035c|M.abscessus_ATCC_199 AAALADGKTVNTEVTASPTIFLPGSNATLENYNRASCGGGDTTSLKEAFA
TH_0586|M.thermoresistible__bu ATAIEDGLTTPDEVHHVPGSIHMGGVTVGDAWEHGVMPYTTTG----IFG
::*: * :: . .. : : : * *
Mflv_0810|M.gilvum_PYR-GCK HSCNTAFVQLGIDTG-ADALRSTAVGFGVDTAPPAIPLQVAESTIGPIVD
Mvan_0025|M.vanbaalenii_PYR-1 HSCNTAFVQLGIDTG-ADPLRSTAGDFGVGAVPPAIPLQVAESTVGPIAD
MSMEG_0031|M.smegmatis_MC2_155 KSCNTAFVELGLSTG-TDKLKAMAQAFGLDTPPPAIPLQVAESTTGPIVD
Mb0016c|M.bovis_AF2122/97 KSCNTAFVQLGIRTG-ADALRSMARAFGLDSPPRPTPLQVAESTVGPIPD
Rv0016c|M.tuberculosis_H37Rv KSCNTAFVQLGIRTG-ADALRSMARAFGLDSPPRPTPLQVAESTVGPIPD
MMAR_0018|M.marinum_M KSCNTAFVQLGISTG-AAALRSMAHAFGLDTPPSAIPLQVAESTVGPIPD
MUL_0020|M.ulcerans_Agy99 KSCNTAFVQLGISTG-AAALRSMAHAFGLDTPPSAIPLQVAESTVGPIPD
MAV_0020|M.avium_104 LSCNTAFVQLGILTG-ADALRSMARSFGLDSTPSVIPLQVAESTIGIIPD
MLBr_00018|M.leprae_Br4923 MSCNTAFVQLGLLIG-ADALRSMAHSFGLDSNPNVIPLQVAESTVGVIPD
MAB_0035c|M.abscessus_ATCC_199 RSCNTAFAKLGAEQLKRDKLVKAAQAFGLDRELDSIPLQVATSTVGPMSD
TH_0586|M.thermoresistible__bu KSSNVGTLMLAQRIG-PERYYEMLRKFGLGQRTGVGLPGESAGLLPPIDQ
*.*.. *. **:. : . : :
Mflv_0810|M.gilvum_PYR-GCK DAALGMTSIGQ-RDVAMTPLQNAMVAATVANNGVTMTPYLVESLKGPDLA
Mvan_0025|M.vanbaalenii_PYR-1 DAALGMSSIGQ-RDVALTPLQNAMIAAAVANKGVMMRPYLVESLKGPDLA
MSMEG_0031|M.smegmatis_MC2_155 AAALGMSSIGQ-RDVALTPLQNAQVAATIANDGIAMRPYLVESLKGPDLA
Mb0016c|M.bovis_AF2122/97 SAALGMTSIGQ-KDVALTPLANAEIAATIANGGITMRPYLVGSLKGPDLA
Rv0016c|M.tuberculosis_H37Rv SAALGMTSIGQ-KDVALTPLANAEIAATIANGGITMRPYLVGSLKGPDLA
MMAR_0018|M.marinum_M AAALGMSSIGQ-KDVALTPLQNAQIAATIANGGVVMRPYLVDSLKGPDLA
MUL_0020|M.ulcerans_Agy99 AAALGMSSIGQ-KDVALTPLQNAQIAATIANGGVVMRPYLVDGLKGPDLA
MAV_0020|M.avium_104 AAALGMSSIGQ-KDVALTPLQNAEIAATIANGGVTMQPYLVDSLKGPDLT
MLBr_00018|M.leprae_Br4923 AAALGMSSIGQ-RDVALTPLQNAQIAATIANAGVTMQPYLIDNLKGPDLA
MAB_0035c|M.abscessus_ATCC_199 DAAVAMSSIGQ-KDVAMTPLQNAMVAAAVANKGVLMQPHLVDSLKGPDLS
TH_0586|M.thermoresistible__bu WSGSTFSNLPIGQGLSMTLLQMAAMYQTIANDGVRVPPRIVKATIAPDGT
:. ::.: :.:::* * * : ::** *: : * :: .** :
Mflv_0810|M.gilvum_PYR-GCK NIATTSPTEERRAVPEKVADTLTDLMVAAEQV--------TQQKGAIAGV
Mvan_0025|M.vanbaalenii_PYR-1 NIATTAPTEERRAVTEQVADTLTDLMVAAEQV--------TQQKGAIAGV
MSMEG_0031|M.smegmatis_MC2_155 TISTTTPEQERRAVSPQVAATLTDLMVAAEQV--------TQQKGAIAGV
Mb0016c|M.bovis_AF2122/97 NISTTVGYQQRRAVSPQVAAKLTELMVGAEKV--------AQQKGAIPGV
Rv0016c|M.tuberculosis_H37Rv NISTTVGYQQRRAVSPQVAAKLTELMVGAEKV--------AQQKGAIPGV
MMAR_0018|M.marinum_M NISTTTPHQQRRAVSPQVAAKLTELMVGAEQV--------AQQKGAIPGV
MUL_0020|M.ulcerans_Agy99 NISTTAPHQQRRAVSPQVAAKLTELMVGAEQV--------AQQKGAIPGV
MAV_0020|M.avium_104 TISTTTPYEQRRAVSPQVAAKLTELMVGAEKV--------AQQKGAIPGV
MLBr_00018|M.leprae_Br4923 NIRTTTSYQQHRAVSPQIAATLTELMVGAEKV--------TQQKGAIPGV
MAB_0035c|M.abscessus_ATCC_199 NLSTTTPDDIGQAVTPAIANTLTELMIASEQR--------TAQTGALPGV
TH_0586|M.thermoresistible__bu RTEEPRP-EGVRVVSAETARIVRDMFRAVVQRDPRGIQQGTGPQAAVEGF
. : :.*. * : ::: . : : .*: *.
Mflv_0810|M.gilvum_PYR-GCK QIASKTGTAEHGTDPR-NTPPHAWYIAFAPAQ-----------APTVAVA
Mvan_0025|M.vanbaalenii_PYR-1 QIASKTGTAEHGTDPR-NTPPHAWYIAFAPAQ-----------APKVAVA
MSMEG_0031|M.smegmatis_MC2_155 QIASKTGTAEHGTDPR-NTPPHAWYIAFAPAQ-----------DPKVAVA
Mb0016c|M.bovis_AF2122/97 QIASKTGTAEHGTDPR-HTPPHAWYIAFAPAQ-----------APKVAVA
Rv0016c|M.tuberculosis_H37Rv QIASKTGTAEHGTDPR-HTPPHAWYIAFAPAQ-----------APKVAVA
MMAR_0018|M.marinum_M QIASKTGTAEHGTDPR-HTPPHAWYIAFAPAQ-----------APKVAVA
MUL_0020|M.ulcerans_Agy99 QIASKTGTAEHGTDPR-HTPPHAWYIAFAPAQ-----------APKVAVA
MAV_0020|M.avium_104 QIASKTGTAEHGSDPR-HTPPHAWYIAFAPAQ-----------TPKVAVA
MLBr_00018|M.leprae_Br4923 QIASKTGTAEHGTDPR-HTPPHAWYIAFAPVQ-----------APKVAVA
MAB_0035c|M.abscessus_ATCC_199 QIASKTGTAEHGTDPR-NTPPHAWYIAFAPAQGTTTDPQATAVTPKVAIA
TH_0586|M.thermoresistible__bu QIAGKTGTAQQINPACGCYYSDVYWITFAGIATAD--------NPRYVIG
***.*****:: . . ...::*:** * .:.
Mflv_0810|M.gilvum_PYR-GCK VLVENGGDR-LGATGGALAAPIGRATIAAALRGAS---------------
Mvan_0025|M.vanbaalenii_PYR-1 VLVENGGNR-LSATGGALAAPIGRATIAAALREAS---------------
MSMEG_0031|M.smegmatis_MC2_155 VLVEDGGDR-LSATGGALAAPIGRATIAAALREGS---------------
Mb0016c|M.bovis_AF2122/97 VLVENGADR-LSATGGALAAPIGRAVIEAALQGEP---------------
Rv0016c|M.tuberculosis_H37Rv VLVENGADR-LSATGGALAAPIGRAVIEAALQGEP---------------
MMAR_0018|M.marinum_M VLVENGADR-LSATGGALAAPIGRAVIEAALQGEP---------------
MUL_0020|M.ulcerans_Agy99 VLVENGADR-LSATGGALAAPIGQAVIEAALQGEP---------------
MAV_0020|M.avium_104 VLVENGADR-LSATGGALAAPIGRAVIEAALQGGP---------------
MLBr_00018|M.leprae_Br4923 VLVEKGADS-LFATGGALAAPIGRAVIEAALQGGA---------------
MAB_0035c|M.abscessus_ATCC_199 VLVEDGGDRALAATGGSVAAPIGRQVIAAALQGAS---------------
TH_0586|M.thermoresistible__bu VMMD-APHRAADGSPGSSAAPLFHNIASWLLRRENVPLSPDPGPPLLLQA
*::: . . .: *: ***: : *:
Mflv_0810|M.gilvum_PYR-GCK -
Mvan_0025|M.vanbaalenii_PYR-1 -
MSMEG_0031|M.smegmatis_MC2_155 -
Mb0016c|M.bovis_AF2122/97 -
Rv0016c|M.tuberculosis_H37Rv -
MMAR_0018|M.marinum_M -
MUL_0020|M.ulcerans_Agy99 -
MAV_0020|M.avium_104 -
MLBr_00018|M.leprae_Br4923 -
MAB_0035c|M.abscessus_ATCC_199 -
TH_0586|M.thermoresistible__bu T