For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPAQREPKVVVLGGGSWGTTVASICARRGPTLQWVRSEETASDINDNHRNSKYLGDDVELAHTLKATTD FSEAAECADVIVMGVPSHGFRGVLTELAKELRPWVPVVSLVKGLEQGTNYRMSQIVDEVLPGHPAGILAG PNIAREVADGYAAAAVLAMPDQHLAANLANLFRTKRFRTYTTDDVVGVEIAGALKNVYAIAVGMGYSLGI GENTRAMVMARAVNEMSKMGVAMGGHRDTFAGLAGMGDLIVTCTSQRSRNRHVGEQLGQGKTIEEIIAAM NQVAEGVKAASVIMEFAEKYGISMPIAREVDGVVNHGSTVEQAYHGLVAEKPGHEVHGSGF
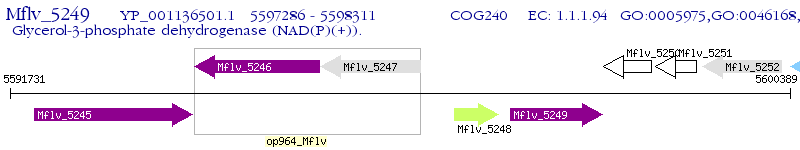
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5249 | - | - | 100% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4221 | gpsA | 1e-59 | 39.68% (315) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0579c | gpsA | 1e-166 | 82.99% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. tuberculosis H37Rv | Rv0564c | gpsA | 1e-166 | 82.99% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_01679 | gpsA | 4e-57 | 37.88% (330) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3915 | - | 1e-150 | 75.15% (338) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. marinum M | MMAR_0915 | gpdA1 | 1e-169 | 84.16% (341) | glycerol-3-phosphate dehydrogenase, GpdA1 |
| M. avium 104 | MAV_4577 | - | 1e-168 | 84.46% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1140 | - | 1e-172 | 87.68% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. thermoresistible (build 8) | TH_2464 | - | 1e-170 | 86.80% (341) | PUTATIVE Glycerol-3-phosphate dehydrogenase (NAD(P)(+)) |
| M. ulcerans Agy99 | MUL_0667 | gpdA1 | 1e-168 | 83.58% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0995 | - | 0.0 | 93.55% (341) | NAD(P)H-dependent glycerol-3-phosphate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0579c|M.bovis_AF2122/97 MAAN----KREPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSAVTA
Rv0564c|M.tuberculosis_H37Rv MAAN----KREPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSAVTA
MMAR_0915|M.marinum_M MAPA----TREPNVVVLGGGSWGTTVASICARRG-P---TLQWVRSEATA
MUL_0667|M.ulcerans_Agy99 MAPA----TREPNVVVLGGGSWGTTVASICARRG-P---TLQWVRSEATA
MAV_4577|M.avium_104 MAAQ----MREPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSRETA
Mflv_5249|M.gilvum_PYR-GCK MAPA----QREPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSEETA
Mvan_0995|M.vanbaalenii_PYR-1 MAPA----QREPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSAETA
MSMEG_1140|M.smegmatis_MC2_155 MAAS----LRVPKVVVLGGGSWGTTVASICARRG-P---TLQWVRSEETA
TH_2464|M.thermoresistible__bu MAAA----KRAPTVVVLGGGSWGTTVANICARRG-P---TLQWVRSEETA
MAB_3915|M.abscessus_ATCC_1997 -MAA----LREPQVVVLGGGSWGTTVASIVARRS-P---TLQWARSPETV
MLBr_01679|M.leprae_Br4923 MEMAGIVVRSESAVAVMGAGAWGTALAKVLIDAGGPEAGVVLWARRPDVA
. *.*:*.*:***::*.: . * .: *.* ..
Mb0579c|M.bovis_AF2122/97 QDINDNHRNSRYLGNDVVLSDTLRATTDFTEAANCADVVVMGVPSHGFRG
Rv0564c|M.tuberculosis_H37Rv QDINDNHRNSRYLGNDVVLSDTLRATTDFTEAANCADVVVMGVPSHGFRG
MMAR_0915|M.marinum_M KDINDNHRNSRYLGNDVVLSETLRATTDFSEAANCADVVVMGVPSHGFRG
MUL_0667|M.ulcerans_Agy99 KDINDNHRNSRYLGNDVVLSETLRATTDFSEAANCADVVVMGVPSHGFRG
MAV_4577|M.avium_104 DDINENHRNSRYLGNDVVLSDTLRATTDFCEAANTADVVVMGVPSHGFRG
Mflv_5249|M.gilvum_PYR-GCK SDINDNHRNSKYLGDDVELAHTLKATTDFSEAAECADVIVMGVPSHGFRG
Mvan_0995|M.vanbaalenii_PYR-1 ADINENHRNSKYLGDEVELAHTLRSTTDFTEAAECADVIVMGVPSHGFRG
MSMEG_1140|M.smegmatis_MC2_155 KDINENHRNSRYLGDDVVLPDSLTATNDFSEAAACADVIVMGVPSHGFRG
TH_2464|M.thermoresistible__bu RDINENHRNSRYLGEDVVLTPDLRATTDFSEAANSADVVVMGVPSHGFRA
MAB_3915|M.abscessus_ATCC_1997 ADINERHRNSRYLSDEVELTESLRATSDLHEAVEQADVVIMGVPSHSFRE
MLBr_01679|M.leprae_Br4923 ERINTTRCNRAYLP-GTLLPPGIRATADPADALRGASTVLLGVPAQRMRA
** : * ** . *. : :* * :* *..:::***:: :*
Mb0579c|M.bovis_AF2122/97 VLVELSKELRPWVPVVSLVKGLEQGTNMRMSQIIEEVLPGHPAG--ILAG
Rv0564c|M.tuberculosis_H37Rv VLVELSKELRPWVPVVSLVKGLEQGTNMRMSQIIEEVLPGHPAG--ILAG
MMAR_0915|M.marinum_M VLTELSKELRPWVPVVSLVKGLEQGTNKRMSQIIEEILPGHPAG--ILAG
MUL_0667|M.ulcerans_Agy99 VLTELSKELRPWVPVVSLVKGLEQGTNKRMSQIIEEILPGHPAG--ILAG
MAV_4577|M.avium_104 VLTELARELRPWVPVVSLVKGLEQGTNMRMSQIVEEVLPGHPAG--ILAG
Mflv_5249|M.gilvum_PYR-GCK VLTELAKELRPWVPVVSLVKGLEQGTNYRMSQIVDEVLPGHPAG--ILAG
Mvan_0995|M.vanbaalenii_PYR-1 VLTELAAHLRPWVPVVSLVKGLEQGTNYRMSQIVNEILPGHPAG--ILAG
MSMEG_1140|M.smegmatis_MC2_155 VLQELARELRPWVPVVSLVKGLEQGTNLRMSQIVDEVLPGHPAG--ILAG
TH_2464|M.thermoresistible__bu VLTELARELRPWVPVVSLVKGLEQGTNLRMSQIVNEVLPGHPAG--ILAG
MAB_3915|M.abscessus_ATCC_1997 VLTEIGQSLRPWVPIVSLVKGLEQGSRMRMSQIIEEVLPGHPAG--ILAG
MLBr_01679|M.leprae_Br4923 NLERWGGLVADGATLVSLAKGIELGTLMRMSQVIVSVTGVDPAQVAVLSG
* . . : ..:***.**:* *: ****:: .: .** :*:*
Mb0579c|M.bovis_AF2122/97 PNIAREVAEGYAAAAVLAMPDQHLATRLSAMFRTRRFRVYTTDDVVGVET
Rv0564c|M.tuberculosis_H37Rv PNIAREVAEGYAAAAVLAMPDQHLATRLSAMFRTRRFRVYTTDDVVGVET
MMAR_0915|M.marinum_M PNIAREVAEGYAAAAVLAMPDQHLATQLSALFRTRRFRVYTTDDVIGVEM
MUL_0667|M.ulcerans_Agy99 PNIAREVAEGYAAAAVLAMPDQHLATQLSALFRTRRFRVYTTDDVIGVEM
MAV_4577|M.avium_104 PNIAREVAEGYAAAAVLAMPDQHLATRLSGLFRTRRFRVYTTDDVVGAEM
Mflv_5249|M.gilvum_PYR-GCK PNIAREVADGYAAAAVLAMPDQHLAANLANLFRTKRFRTYTTDDVVGVEI
Mvan_0995|M.vanbaalenii_PYR-1 PNIAREVAEGYAAAAVLAMPDQSLAANLAQLFRTKRFRTYTTDDVVGVEM
MSMEG_1140|M.smegmatis_MC2_155 PNIAREVAEGYAAAAVLAMPDQHLAAKLAELFRTKRFRTYTTDDVTGVEM
TH_2464|M.thermoresistible__bu PNIAREVAEGYAAAAVLAMPDQEMAAYLARMFRTKRFRTYTTDDVVGVEM
MAB_3915|M.abscessus_ATCC_1997 PNIAKEVARGYAAAGVVAMPDQHQAARLGELFRTSRFRVYSTNDVVGVEM
MLBr_01679|M.leprae_Br4923 PNLASEIAQCQPAATVIACSDLGRAVALQRMLSSGYFRPYTNSDVVGTEI
**:* *:* .** *:* .* *. * :: : ** *:..** *.*
Mb0579c|M.bovis_AF2122/97 AGALKNVFAIAVGMGYSLGIGENTRALVIARALREMTKLGVAMGGKSETF
Rv0564c|M.tuberculosis_H37Rv AGALKNVFAIAVGMGYSLGIGENTRALVIARALREMTKLGVAMGGKSETF
MMAR_0915|M.marinum_M AGALKNVFAIAVGMGYSLGIGENTRALVIARSLREMTKLGVAMGGDTETF
MUL_0667|M.ulcerans_Agy99 AGALKNVFAIAVGMGYSLGIGENPRALVIARSLREMTKLGVVMGGDTETF
MAV_4577|M.avium_104 AGALKNVFAIAVGMGYSLGIGENTRALVIARALREMTKLGVAMGGSPDTF
Mflv_5249|M.gilvum_PYR-GCK AGALKNVYAIAVGMGYSLGIGENTRAMVMARAVNEMSKMGVAMGGHRDTF
Mvan_0995|M.vanbaalenii_PYR-1 AGALKNVYAIAVGMGYSLGIGENTRAMVMARAVAEMSKLGVAMGGNRDTF
MSMEG_1140|M.smegmatis_MC2_155 AGALKNVYAIAVGMGYSLGIGENTRAMVMARAVREMAKLGEAAGGHRDTF
TH_2464|M.thermoresistible__bu AGALKNVYAIAVGMGYSLGIGENTRAMVMARALREMSKLGEAMGGQRDTF
MAB_3915|M.abscessus_ATCC_1997 AGALKNVFAIASGMGYAIGIGENTRAMVIARALREMTKLGVAMGGNPETF
MLBr_01679|M.leprae_Br4923 GGVCKNVIALACGMAAGVGFGENTAATIITRGLAEIIRLGMALGAQVTTL
.*. *** *:* **. .:*:***. * :::*.: *: ::* . *. *:
Mb0579c|M.bovis_AF2122/97 PGLAGLGDLIVTCTSQRSRNRHVGEQLGAGKPIDEIIASMS-------QV
Rv0564c|M.tuberculosis_H37Rv PGLAGLGDLIVTCTSQRSRNRHVGEQLGAGKPIDEIIASMS-------QV
MMAR_0915|M.marinum_M PGLAGLGDLIVTCTSQRSRNRHVGEQLGAGKPIDEIIASMN-------QV
MUL_0667|M.ulcerans_Agy99 PGLAGLGDLIVTCTSQRSRNRHVGEQLGAGKPIDEIIASMN-------QV
MAV_4577|M.avium_104 PGLAGLGDLIVTCTSQRSRNRHVGEQLGAGKPIDEIIASMN-------QV
Mflv_5249|M.gilvum_PYR-GCK AGLAGMGDLIVTCTSQRSRNRHVGEQLGQGKTIEEIIAAMN-------QV
Mvan_0995|M.vanbaalenii_PYR-1 AGLAGMGDLIVTCTSQRSRNRHVGEQLGQGKTVEEIIAAMN-------QV
MSMEG_1140|M.smegmatis_MC2_155 AGLAGMGDLIVTCTSQRSRNRHVGEQLGAGKKIDEIIASMN-------QV
TH_2464|M.thermoresistible__bu AGVAGMGDLIVTCTSKRSRNRHVGEQLGAGKPIDEIIASMD-------QV
MAB_3915|M.abscessus_ATCC_1997 PGLAGLGDLIVTCTSSSSRNRHVGEEIGKGKTIDEIITSMN-------QV
MLBr_01679|M.leprae_Br4923 AGLAGVGDLVATCTSPHSRNRSLGERLGRGEIMQSILHGMDGSDGGDGYV
.*:**:***:.**** **** :**.:* *: ::.*: .*. *
Mb0579c|M.bovis_AF2122/97 AEGVKAAGVVMEFANEFGLNMPIAREVDAVINHGSTVEQAYRGLIAEVPG
Rv0564c|M.tuberculosis_H37Rv AEGVKAAGVVMEFANEFGLNMPIAREVDAVINHGSTVEQAYRGLIAEVPG
MMAR_0915|M.marinum_M AEGVKAASVIMEFANEFGLPMPIAREVDAVINHGSTVEQAYHGLAAEVPG
MUL_0667|M.ulcerans_Agy99 AEGVKAASVIMEFANEFGLPMPIAREVDAVINHGSTVEQAYHGLAAEVPG
MAV_4577|M.avium_104 AEGVKAASVIMEFANEFGLTMPIAREVDAVINHGSTVEQAYRGLIAEVPG
Mflv_5249|M.gilvum_PYR-GCK AEGVKAASVIMEFAEKYGISMPIAREVDGVVNHGSTVEQAYHGLVAEKPG
Mvan_0995|M.vanbaalenii_PYR-1 AEGVKAASVIMEFADKCGISMPIAREVDGVVNHGSTVEQAYRGLVAEKPG
MSMEG_1140|M.smegmatis_MC2_155 AEGVKAASVIMEFADQYGLNMPIAREVDAVINHGSSVEQAYRGLMAEKTG
TH_2464|M.thermoresistible__bu AEGVKAASVIMEFADEYGISMPIAREVDAVVNHGSTVEQAYRGLMAEKPG
MAB_3915|M.abscessus_ATCC_1997 AEGVKASSVVMTLADEYGIQMPIAREVDGVINQGSTVEQAYRGLMAETPG
MLBr_01679|M.leprae_Br4923 VEGVTSCASVLALASSYDVEMPLTDAVHRVCHKGLSVEKAMALLLGRSTK
.***.:.. :: :*.. .: **:: *. * ::* :**:* * .. .
Mb0579c|M.bovis_AF2122/97 HEVHGSGF
Rv0564c|M.tuberculosis_H37Rv HEVHGSGF
MMAR_0915|M.marinum_M HEVYGAGF
MUL_0667|M.ulcerans_Agy99 HEVYGAGF
MAV_4577|M.avium_104 HEVHGSGF
Mflv_5249|M.gilvum_PYR-GCK HEVHGSGF
Mvan_0995|M.vanbaalenii_PYR-1 HEVHGSGF
MSMEG_1140|M.smegmatis_MC2_155 HEVHSSGF
TH_2464|M.thermoresistible__bu HEVHGSGF
MAB_3915|M.abscessus_ATCC_1997 HEVHGTGF
MLBr_01679|M.leprae_Br4923 SE------
*