For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPGSVTAVPVFGGDRFDIVDQYGRQPAELTVLAADPRAVADAPPDTAATVLRSLIPGSDENGYAAQRIL SLLARHRVDQQEAVATRLFGHHSPAGSRATFTVDADAVVLVAAPASPMHPHSDDPNPPSEVRVAVYRADP GRPATPELPPPLAEPLLDLRIDAETAASYEVREGQFIQIIDVAGRQCSDFLAFDARGLADGREYGLDATT TRTIGGAAYPRPGLRGKFYDARARPLVEVVRDTVGRHDTFALACTAKYYADLGYLGHVNCTDNFNGALAR FGVDRRQGWPALNLFYNTAFDAAHQLISDEPWSCAGDYVLFRASADLVCASSACPDDIDPANGWVPTDIH VRVYDSSRRFSVAVGHRLTPDSEPVLTKSTAFARRTGALTTDFTEYNGYWLPNRYANHGPHDEYWACREH VAVMDLSALRKFEVLGPDAEALLQATLTRDIRRLSRGQVVYSAMCSESGGVVDDCTVLRLGDNNFRFIGG DPHGGVWLRTQAERLGLHQVWIKDSTDQMHNLAVQGPASRAVLDGLIWTPPGQPALRDLGWFRFLTGRLD GPDGAPLLVSRTGYTGELGYEVWVHPGDAETLWDRLWDAGRPHGLTPLGLEALEMVRIEAGLAAGGHEFD DQTDPFEAGIGFTVPLKTKTDDFVGRAALVERKAHPQRTLVGLRLDSNETTAHGDCVHLGRTQVGVVTSA TRSPLLGASIALCRIAVQHSEPGTRVEIGKLDGHLKRIPATVTTIPFYDSDKTRPRS
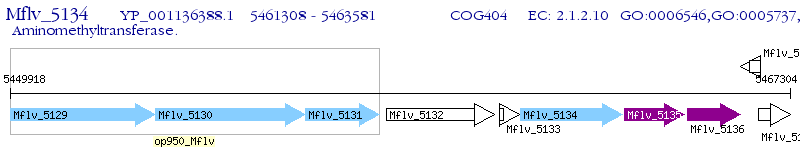
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5134 | - | - | 100% (757) | aminomethyltransferase |
| M. gilvum PYR-GCK | Mflv_2939 | gcvT | 9e-36 | 33.24% (364) | glycine cleavage system aminomethyltransferase T |
| M. gilvum PYR-GCK | Mflv_3080 | - | 2e-32 | 32.59% (313) | FAD dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2234c | gcvT | 1e-33 | 35.22% (335) | glycine cleavage system aminomethyltransferase T |
| M. tuberculosis H37Rv | Rv2211c | gcvT | 1e-33 | 35.22% (335) | glycine cleavage system aminomethyltransferase T |
| M. leprae Br4923 | MLBr_00865 | gcvT | 1e-32 | 33.74% (329) | glycine cleavage system aminomethyltransferase T |
| M. abscessus ATCC 19977 | MAB_1949 | gcvT | 2e-31 | 31.88% (367) | glycine cleavage system aminomethyltransferase T |
| M. marinum M | MMAR_0210 | gcvT | 3e-45 | 32.38% (383) | aminomethyltransferase GcvT_1ne |
| M. avium 104 | MAV_2280 | gcvT | 9e-33 | 33.84% (328) | glycine cleavage system aminomethyltransferase T |
| M. smegmatis MC2 155 | MSMEG_4278 | gcvT | 5e-34 | 34.62% (338) | glycine cleavage system aminomethyltransferase T |
| M. thermoresistible (build 8) | TH_1298 | gcvT | 4e-39 | 35.85% (371) | Probable aminomethyltransferase GcvT (Glycine cleavage |
| M. ulcerans Agy99 | MUL_3571 | gcvT | 9e-34 | 33.24% (373) | glycine cleavage system aminomethyltransferase T |
| M. vanbaalenii PYR-1 | Mvan_1202 | - | 0.0 | 80.59% (752) | glycine cleavage T protein (aminomethyl transferase) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK MAPGSVTAVPVFGGDRFDIVDQYGRQPAELTVLAADPRAVADAPPDTAAT
Mvan_1202|M.vanbaalenii_PYR-1 -----MTAVRVFGGDRLDVVDHYGRQPAELTVLATDPRAVVGTRPDAPAT
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK VLRSLIPGSDENGYAAQRILSLLARHRVDQQEAVATRLFGHHSPAGSRAT
Mvan_1202|M.vanbaalenii_PYR-1 VLRHLIPGPDENGYAAARILSLLSDHHVHQQDSTATRLFDGPSPAGARVS
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK FTVDADAVVLVAAPASPMHPHSDDPNPPSEVRVAVYRADPGRPATPELPP
Mvan_1202|M.vanbaalenii_PYR-1 FAVDADAVVLVAAPAAAMDLTGPEPNPPSEVRVEVHRADPRRPQVRELPA
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK PLAEPLLDLRIDAETAASYEVREGQFIQIIDVAGRQCSDFLAFDARGLAD
Mvan_1202|M.vanbaalenii_PYR-1 PLAEPLWETRVDAATASSYEVGAGQFIQIIDVAGQQCSDFLAFDARALGD
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK GREYGLDATTTRTIGGAAYPRPGLRGKFYDARARPLVEVVRDTVGRHDTF
Mvan_1202|M.vanbaalenii_PYR-1 GAEFGLDATTTRTIGGGAYPRPGLFGKFFDSRARPLVEVVRDTVGRHDTF
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK ALACTAKYYADLGYLGHVNCTDNFNGALARFGVDRRQGWPALNLFYNTAF
Mvan_1202|M.vanbaalenii_PYR-1 ALACTAKYYADLGYPGHVNCTDNFNATLSRFGVQPRRGWPALNLFYNTAF
Mb2234c|M.bovis_AF2122/97 --------------------------------------------------
Rv2211c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3571|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00865|M.leprae_Br4923 --------------------------------------------------
MAV_2280|M.avium_104 --------------------------------------------------
MSMEG_4278|M.smegmatis_MC2_155 --------------------------------------------------
TH_1298|M.thermoresistible__bu --------------------------------------------------
MAB_1949|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0210|M.marinum_M --------------------------------------------------
Mflv_5134|M.gilvum_PYR-GCK DAAHQLISDEPWSCAGDYVLFRASADLVCASSACPDDIDPANGWVPTDIH
Mvan_1202|M.vanbaalenii_PYR-1 DAQHQLVSDEPWSRPGDYVLLRACSDLVCASSACPDDIDAANGWLPTDIH
Mb2234c|M.bovis_AF2122/97 --------MCQQGRPLGWDAVSDVPELIHGPLEDRHRELGASFAEFGGWL
Rv2211c|M.tuberculosis_H37Rv --------MCQQGRPLGWDAVSDVPELIHGPLEDRHRELGASFAEFGGWL
MUL_3571|M.ulcerans_Agy99 --------------------MSDAPELMQGPLEDRHRALGASFAEFGGWL
MLBr_00865|M.leprae_Br4923 --------------------MTDAPELLKGPLEDRHRELGANFAEFGGWL
MAV_2280|M.avium_104 --------------------MSNEADLLHGPLEDRHRDLGASFAEFGGWL
MSMEG_4278|M.smegmatis_MC2_155 --------------------MSD--ELLQGPLEDRHRELGAAFAEFGGWL
TH_1298|M.thermoresistible__bu --------------------VTGVSDIQHGPLEERHRQLGANFAEFGGWL
MAB_1949|M.abscessus_ATCC_1997 -----------------------MTDLLLGPLHDRHAAQGATFAEFGGWN
MMAR_0210|M.marinum_M --------------------MTTSRTTPATPLLQQHHELGARFTDFAGWQ
Mflv_5134|M.gilvum_PYR-GCK VRVYDSSRRFSVAVGHRLTPDSEPVLTKSTAFARRTGALTTDFTEYNGYW
Mvan_1202|M.vanbaalenii_PYR-1 VRVYDSTRRFSVAVGHRLTPDSEPVLTKPTAFANRTGALTSNFTEYKGYW
.: : : *::: *:
Mb2234c|M.bovis_AF2122/97 MPVSYA--GTVSEHNATRTAVGLFDVSHLGKALVRGPGAAQFVNSALTND
Rv2211c|M.tuberculosis_H37Rv MPVSYA--GTVSEHNATRTAVGLFDVSHLGKALVRGPGAAQFVNSALTND
MUL_3571|M.ulcerans_Agy99 MPVSYT--GTVSEHNATRNAAGLFDVSHLGKALVRGPGAAEFVNSALTND
MLBr_00865|M.leprae_Br4923 MPVSYA--GTVSEHSATRNAVGLFDVSHLGKALVRGPGAAQFVNSVLTND
MAV_2280|M.avium_104 MPVSYA--GTVSEHNATRNAVGLFDVSHLGKALVRGTGAARFVNSALTND
MSMEG_4278|M.smegmatis_MC2_155 MPVSYA--GTVAEHNATRSTVGLFDVSHLGKALVKGPGAAAYVNSALTND
TH_1298|M.thermoresistible__bu MPVSYA--GTVSEHTATRTTVGLFDVSHLGKARVRGPGAADFVNSAFTND
MAB_1949|M.abscessus_ATCC_1997 MPVSYA--GTVGEHTATRAAVGLFDVSHLGKALVRGPGAAAFVNACFTND
MMAR_0210|M.marinum_M MPVRYG--SELTEHHTVRSAAGLFDLSHMGEIEVLGPGAAAALDYALVGA
Mflv_5134|M.gilvum_PYR-GCK LPNRYANHGPHDEYWACREHVAVMDLSALRKFEVLGPDAEALLQATLTRD
Mvan_1202|M.vanbaalenii_PYR-1 LPNSFDNHGPHQEYWACRERVAVMDLSALRKFEVLGPDAEELLQATLTRD
:* : . *: : * ..::*:* : : * *..* :: :.
Mb2234c|M.bovis_AF2122/97 LGRIGPGKAQYTLCCTESGGVIDDLIAYYVSDDEIFLVPNAANTAAVVGA
Rv2211c|M.tuberculosis_H37Rv LGRIGPGKAQYTLCCTESGGVIDDLIAYYVSDDEIFLVPNAANTAAVVGA
MUL_3571|M.ulcerans_Agy99 LRRIGPGKAQYTLCCNESGGVIDDLIAYYVADDEIFLVPNAANTAAVVAA
MLBr_00865|M.leprae_Br4923 LGRIRPGKAQYTLCCSESGGVIDDLIAYYVDDDEIFLVSNAANTAAVVDA
MAV_2280|M.avium_104 LNRIGPGKAQYTLCCNESGGVIDDLIAYYVDDDEIFLVPNAANTAAVVEA
MSMEG_4278|M.smegmatis_MC2_155 LGKIGPGKAQYTLCCTESGGVIDDLIAYYVSDDEIFLVPNAANTATVVEE
TH_1298|M.thermoresistible__bu LRRIGPGSAQYTLCCTESGGVVDDLIAYYISDDEIFLVPNAANNATVVDL
MAB_1949|M.abscessus_ATCC_1997 LNKVGPGKAQYTLCCTETGGVIDDLIAYYVSDDEIFLVPNAANTSAVVAA
MMAR_0210|M.marinum_M FSGLKVGKAKYSLLCREDGGVIDDLVAYRLAEDNFLVVANAANAAAVYEA
Mflv_5134|M.gilvum_PYR-GCK IRRLSRGQVVYSAMCSESGGVVDDCTVLRLGDNNFRFIGGDPHGGVWLRT
Mvan_1202|M.vanbaalenii_PYR-1 VRRLSRGQVVYSAMCTEAGGVVDDCTVLRLGDNNFRFIGGDPYDGIWLRT
. : *.. *: * * ***:** . : :::: .: . . .
Mb2234c|M.bovis_AF2122/97 LQAAAP-GGLSITNLHRSYAVLAVQGPCSTDVLTALGLP-------TEMD
Rv2211c|M.tuberculosis_H37Rv LQAAAP-GGLSITNLHRSYAVLAVQGPCSTDVLTALGLP-------TEMD
MUL_3571|M.ulcerans_Agy99 LQAAAP-SGLTITDLHRSYAVLAVQGPRSTEVLAALGLP-------TEMD
MLBr_00865|M.leprae_Br4923 LQAVVP-AGLTIINQHRSHAVLAVQGPRSTDVLGELGLP-------TGID
MAV_2280|M.avium_104 LQGAAP-AGVTVSNLHRSYAVLAVQGPRSADVLAELGLP-------SDMD
MSMEG_4278|M.smegmatis_MC2_155 LQRHAP-DGLTITNEHRSYAVLAVQGPKSAEVLDKLGLP-------TDMD
TH_1298|M.thermoresistible__bu LRRHAP-AGVEVIDEHRAHAVFAVQGPRSADVLSGLGLP-------VDMD
MAB_1949|M.abscessus_ATCC_1997 LQEQAP-EGIAVTNQHRDYAVLAVQGPRSADVLQRLGLP-------TDME
MMAR_0210|M.marinum_M LVQRAAGFDATVTDRSQDTALIALQGPVAERVLCKVVAEA-DRATVTELK
Mflv_5134|M.gilvum_PYR-GCK QAERLGLHQVWIKDSTDQMHNLAVQGPASRAVLDGLIWTPPGQPALRDLG
Mvan_1202|M.vanbaalenii_PYR-1 QAERLGLGQVWIKDSSDHMHNLAVQGPSSRELLCELIWSPPGQPALRDLG
: : :*:*** : :* : :
Mb2234c|M.bovis_AF2122/97 YMGYADASYSG---VPVRVCRTGYTGEHGYELLPPWESAGVVFDALLAAV
Rv2211c|M.tuberculosis_H37Rv YMGYADASYSG---VPVRVCRTGYTGEHGYELLPPWESAGVVFDALLAAV
MUL_3571|M.ulcerans_Agy99 YMGYADSSYNG---VSVRVCRTGYTGEHGYELLPPWESAGVVFDALAAAV
MLBr_00865|M.leprae_Br4923 YMGYVDASYAG---VPVRVCRTGYTGEQGYELLPPWESADVVFDALVAAV
MAV_2280|M.avium_104 YMAYADTSFRQ---VPVRVCRTGYTGEHGYELLPPWESAGVVFDALAAAV
MSMEG_4278|M.smegmatis_MC2_155 YMGYADAEFDG---VFVRVCRTGYTGEHGYELLPAWDRAGVVFDALVAAV
TH_1298|M.thermoresistible__bu YMAFTDATLNG---VEVRVCRTGYTGEHGYEVLPPWESALPVFDALVDAV
MAB_1949|M.abscessus_ATCC_1997 YMAYADATLAG---LPVRVCRTGYTGEHGYELLPSWDDAGAVFDALLPVI
MMAR_0210|M.marinum_M YYAATQIQLGE---IPVLLARTGYTGEDGFELYVDNAHAAPVWHLLLDAG
Mflv_5134|M.gilvum_PYR-GCK WFRFLTGRLDGPDGAPLLVSRTGYTGELGYEVWVHPGDAETLWDRLWDAG
Mvan_1202|M.vanbaalenii_PYR-1 WFRFLIGRLDGPEGPPLLVSRTGYTGELGYELWIHPNDAETLWDRVWEAG
: : :.******* *:*: * ::. : .
Mb2234c|M.bovis_AF2122/97 SAAGGEPAGLGARDTLRTEMGYPLHGHELSLDISPLQARCGWAVGWRKD-
Rv2211c|M.tuberculosis_H37Rv SAAGGEPAGLGARDTLRTEMGYPLHGHELSLDISPLQARCGWAVGWRKD-
MUL_3571|M.ulcerans_Agy99 AEVGGSPAGLGARDTLRTEMGYPLHGHELSLDISPLQARCGWAIGWKKD-
MLBr_00865|M.leprae_Br4923 VDARGEPAGLGARDTLRTEMGYPLYGHELSLDISPLQARCGWAIGWKKD-
MAV_2280|M.avium_104 SQAGGQPAGLGARDTLRTEMGYPLHGHELSPDISPLQARCGWAIGWKKE-
MSMEG_4278|M.smegmatis_MC2_155 RAAGGEPAGLGARDTLRTEMGYPLHGHELSLDISPLQARCGWAVGWKKD-
TH_1298|M.thermoresistible__bu QAAGGQLAGLGARDTLRTEMGYPLHGHELSPDISPLEARCGWAVGWKKE-
MAB_1949|M.abscessus_ATCC_1997 TEAGGQLAGLGARDTLRTEMGYPLHGHELSLDISPVQARAGWAVGWKKD-
MMAR_0210|M.marinum_M RADGVIPAGLACRDTLRLEAGMALYGHELSLETSPYDAGLGRTVRLEK--
Mflv_5134|M.gilvum_PYR-GCK RPHGLTPLGLEALEMVRIEAGLAAGGHEFDDQTDPFEAGIGFTVPLKTKT
Mvan_1202|M.vanbaalenii_PYR-1 QAYGLAPLGLEALELLRVESGLVAAGHEFDEQTDPFEAGIGFTVPLKSKS
** . : :* * * ***:. : .* :* * :: ..
Mb2234c|M.bovis_AF2122/97 -AFFGRAALLAEKAAGPRRLLRGLRMVGRGVLRPGLAVLV--GDETVGVT
Rv2211c|M.tuberculosis_H37Rv -AFFGRAALLAEKAAGPRRLLRGLRMVGRGVLRPGLAVLV--GDETVGVT
MUL_3571|M.ulcerans_Agy99 -AFFGREALLAEKAAGPRRLPRGLRMVGRGVLRPGLTVLN--GSTPVGVT
MLBr_00865|M.leprae_Br4923 -AFLGRDALLAEKAAGPRRLLRGLRMAGRGVLRPGLTVCA--GDIPIGVT
MAV_2280|M.avium_104 -AFFGRDALLAEKEAGPRRLLRGLRMVGRGVLRAGLTVLV--GDTPVGVT
MSMEG_4278|M.smegmatis_MC2_155 -AFWGRDALLAEKADGPKRVLRGLKALGRGVLRADLAVLD--GDTRVGVT
TH_1298|M.thermoresistible__bu -RFWGRDALLAEKQAGPRRLLRGLRAVDRGVLRAGLSVYD--GDTVVGTT
MAB_1949|M.abscessus_ATCC_1997 -AFWGREALTQEKTDGPRRTLRGLRATGRGVLRPDLTVLS--DGQSIGVT
MMAR_0210|M.marinum_M -YFVGRDALAELAAQGPRRTLIGLQGLGKRAARAGYAVYTEDGQRQIGTV
Mflv_5134|M.gilvum_PYR-GCK DDFVGRAALVERKAH-PQRTLVGLRLDSNETTAHGDCVHL--GRTQVGVV
Mvan_1202|M.vanbaalenii_PYR-1 DDFVGRDALIERKAH-PQRALVGLRLDGNEVAAHGDCVHI--GRSQVGVV
* ** ** *:* **: .. . . * . :*..
Mb2234c|M.bovis_AF2122/97 TSGTFSPTLQVGIGLALIDSDAGIEDGQQINVDVRG--RAVECQVVCPPF
Rv2211c|M.tuberculosis_H37Rv TSGTFSPTLQVGIGLALIDSDAGIEDGQQINVDVRG--RAVECQVVCPPF
MUL_3571|M.ulcerans_Agy99 TSGTFSPTLQIGIALALIDTDAGVEDGQQITVDVRG--RAVECEVVRPPF
MLBr_00865|M.leprae_Br4923 TSGTFSPTLQVGVALALIDSEAAVQDGQQIIVDVRG--RAVECEVVRPPF
MAV_2280|M.avium_104 TSGTFSPTLQAGIALALIDTDADVRDGQKVTVDVRG--RAATCEVVRPPF
MSMEG_4278|M.smegmatis_MC2_155 TSGTFSPTLKVGIALALIDTEYDIADGTTVTVDVRG--RAVECEVVKPPF
TH_1298|M.thermoresistible__bu TSGTFSPTLKVGIALALIDTEHGIPDGARVSVDVRG--RRIGCEVVKPPF
MAB_1949|M.abscessus_ATCC_1997 TSGTFSPTLKTGIALALLDTAAQIPDGASVVVDVRG--REIECEVVKPPF
MMAR_0210|M.marinum_M TSGALSPTLGYPIAISYVEAGT-VDTDSIVAVDIRG--SMEHFRVVATPF
Mflv_5134|M.gilvum_PYR-GCK TSATRSPLLGASIALCRIAVQHSEPGTRVEIGKLDGHLKRIPATVTTIPF
Mvan_1202|M.vanbaalenii_PYR-1 TSGIRSPVLGAGIALCRIAVQYTAPGTRVEVGKLDGHRKRLAATVTTVPF
**. ** * :.:. : .: * *. **
Mb2234c|M.bovis_AF2122/97 VAVKTR----
Rv2211c|M.tuberculosis_H37Rv VAVKTR----
MUL_3571|M.ulcerans_Agy99 VEAKTR----
MLBr_00865|M.leprae_Br4923 IEVKTR----
MAV_2280|M.avium_104 VAVKTR----
MSMEG_4278|M.smegmatis_MC2_155 VTPSTR----
TH_1298|M.thermoresistible__bu VEVKTR----
MAB_1949|M.abscessus_ATCC_1997 VDVNVG----
MMAR_0210|M.marinum_M YVRPRPA---
Mflv_5134|M.gilvum_PYR-GCK YDSDKTRPRS
Mvan_1202|M.vanbaalenii_PYR-1 YDPDKIRPRS