For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VFSTHLSAQLGKALPVQVSHRFAASSAVFDDERLVSCAGLVPVMALAEQTGLSSSLADKVRFSCERVKSG AANPSPKLTTLIAGMCTGADSIDDLDVVRSGGMKTLFGEVYAPSTIGTLLREFTFGHARQLESVLSEHVG ALCERVDLLPGADGRVFIDIDSLLRPVYGQAKQGASYGHTKIAGKQILRKGLSPLVTTISTEHCAPVITG ARLRAGKTNSGKGAARMIAQAVATARGAGVTGRILVRGDSAYGNSSVVTACRRAGARFSLVLTKTAAVVA AIADINDTAWIPVCYPAAIPTPVPGSPMPKSPKPPAPLSVPRRLRSRHG
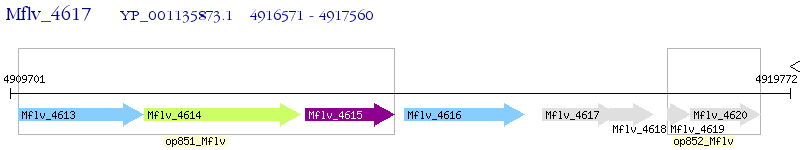
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4617 | - | - | 100% (329) | TnpC protein |
| M. gilvum PYR-GCK | Mflv_3048 | - | 7e-08 | 26.50% (283) | transposase, IS4 family protein |
| M. gilvum PYR-GCK | Mflv_2324 | - | 7e-08 | 26.50% (283) | transposase, IS4 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1481 | - | 2e-90 | 74.66% (221) | TnpC protein |
| M. smegmatis MC2 155 | MSMEG_4127 | - | 1e-129 | 80.41% (291) | TnpC protein |
| M. thermoresistible (build 8) | TH_0489 | - | 1e-111 | 73.02% (278) | TnpC protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5550 | - | 1e-139 | 81.37% (306) | transposase, IS4 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1481|M.avium_104 --------------------------------------------------
TH_0489|M.thermoresistible__bu ---------------------------VFDDDNLVSCAGLVPVMGQAEQT
MSMEG_4127|M.smegmatis_MC2_155 ---------------MELSHRFAASSAVFDDDHLVSIAGLVPVMTLATQT
Mvan_5550|M.vanbaalenii_PYR-1 MFSTHLSTQLRKALPVQVSHKFAASSAVFDDDHLVSCAGLVPVMTLAAQT
Mflv_4617|M.gilvum_PYR-GCK MFSTHLSAQLGKALPVQVSHRFAASSAVFDDERLVSCAGLVPVMALAEQT
MAV_1481|M.avium_104 ----------------------------------MCVGADSIDDLDLVRA
TH_0489|M.thermoresistible__bu GLSDLLAQHVHITETRIKSGAANLAPKLATVIAAMCAGADCIDDVDVLRS
MSMEG_4127|M.smegmatis_MC2_155 GLSALLADKVRISEPRIKSGSANPSPKLTTLIAGMCAGADSIDDLDIVRS
Mvan_5550|M.vanbaalenii_PYR-1 GLAQLLAAKVRITEPRIKSGSANPAPKLTTVVAGMCAGADCIDDLDLVRS
Mflv_4617|M.gilvum_PYR-GCK GLSSSLADKVRFSCERVKSGAANPSPKLTTLIAGMCTGADSIDDLDVVRS
**.***.***:*::*:
MAV_1481|M.avium_104 GGMKSLFGGVYAASTVGTLLREFTFGHARQLESVLRHHLSHLCQRVDLLP
TH_0489|M.thermoresistible__bu GGMKTLFDNVYAPSTIGTLLREFTFGHNRQLESVLRHHLSALCGHVDLLP
MSMEG_4127|M.smegmatis_MC2_155 GGMKTLFDDVYAPSTIGTLLREFTFGHARQLEAVLRAHLAELCQRADLLP
Mvan_5550|M.vanbaalenii_PYR-1 GGMKTLFDGVYAPSTIGTLLREFTFGHARQLESVLREHLAGLCERVQLLP
Mflv_4617|M.gilvum_PYR-GCK GGMKTLFGEVYAPSTIGTLLREFTFGHARQLESVLSEHVGALCERVDLLP
****:**. ***.**:*********** ****:** *:. ** :.:***
MAV_1481|M.avium_104 GSDKGAFIDIDSLLRPVYGHAKQGASYGHTKIAGKQILRKGLSPLVTTIS
TH_0489|M.thermoresistible__bu GAAQRVFVDIDSLLGPVYGHAKQGASYGHTKIAGKQVLRKGLSPLATTIS
MSMEG_4127|M.smegmatis_MC2_155 GIDGRTFVDIDSLLRPVYGHAKQGASYGHTKIAGKQILRKGLSPLITTIS
Mvan_5550|M.vanbaalenii_PYR-1 GSDERAFVDIDSLLRPVYGRAKQGASYGHTKIAGKQILRKGLSPLVTTIS
Mflv_4617|M.gilvum_PYR-GCK GADGRVFIDIDSLLRPVYGQAKQGASYGHTKIAGKQILRKGLSPLVTTIS
* .*:****** ****:****************:******** ****
MAV_1481|M.avium_104 TTGAAPAITGMRLRAGKANSGKGAGRMIAQAIGTARAAGVRGQILVRGDS
TH_0489|M.thermoresistible__bu TDTAAPVIAGMRLQAGKTGSGKGAGRMIAQAIVTARAAGASGQILVRGDS
MSMEG_4127|M.smegmatis_MC2_155 SSTSAPVIAGARLRAGKTNSGKGAARMIAQAVATARAAGVTGPILVRGDS
Mvan_5550|M.vanbaalenii_PYR-1 TETAAPVITGARLRAGKTNSGKGAARMIAQAVATARASGVTGQILVRGDS
Mflv_4617|M.gilvum_PYR-GCK TEHCAPVITGARLRAGKTNSGKGAARMIAQAVATARGAGVTGRILVRGDS
: .**.*:* **:***:.*****.******: ***.:*. * *******
MAV_1481|M.avium_104 AYGTRAVVGACRSHGARFSLVMTRNTAIDRAINSIDESAWTPVRYPGAVR
TH_0489|M.thermoresistible__bu VYGRRAVVKACRRAGAQFSLVWTKNTAVRTAIAAIDEDAWIPVRYPGAVQ
MSMEG_4127|M.smegmatis_MC2_155 AYGNSTVAAACRRAGAQFSLVLTKTPAVTAAIDAISDGAWIPVNYPGAVR
Mvan_5550|M.vanbaalenii_PYR-1 AYGNSAVVAACRRAGARFSLVLTKTAAVAAAIESISESAWVPVKYPGAVR
Mflv_4617|M.gilvum_PYR-GCK AYGNSSVVTACRRAGARFSLVLTKTAAVVAAIADINDTAWIPVCYPAAIP
.** :*. *** **:**** *:..*: ** *.: ** ** **.*:
MAV_1481|M.avium_104 DPDTGDWISDAEVAEIGYTAFASTKDRFTARLVVRRVKDARFRDALFPVW
TH_0489|M.thermoresistible__bu DPDSGKWISDAEVAEIGYTAFASTKDAITARLIVRRVKDARHRDALFPVW
MSMEG_4127|M.smegmatis_MC2_155 DPDTGAWISDAEVAETTYTAFSSTKTPITARLIVRRVKDARFLDALFPVW
Mvan_5550|M.vanbaalenii_PYR-1 DPDTGAWISDAEVAETTYTAFSSTKTPVTARLIVRRVKDARFLDALFPVW
Mflv_4617|M.gilvum_PYR-GCK TPVPG---------------------------------------------
* .*
MAV_1481|M.avium_104 RYHPFFTNTDLPTAEADITHRQHAIIETVFADLIDGPLAHMPSGQFGANS
TH_0489|M.thermoresistible__bu RCHPFLTNSDEPTADADITNRHRAVVETTFADLIDGPLARMPSGHFGANS
MSMEG_4127|M.smegmatis_MC2_155 RYHPFFTDSDEPVDAADITHRRHAIIETVFADLIDGPLAHMPSGRFGANS
Mvan_5550|M.vanbaalenii_PYR-1 RYHPFFTDSDEPVDASDITHRRHAIIETVFADLIDGPLAHMPSGRFGANS
Mflv_4617|M.gilvum_PYR-GCK -----------------------------------SPMPKSP--------
.*:.: *
MAV_1481|M.avium_104 AWVLCAAIAHNLLRAAGVLAGGAHAVARGATLRRKIVNIPARLACPQRRP
TH_0489|M.thermoresistible__bu AWILCAGIAHNLLRGAATLAGEPHIVARGA-MRRKIVNIPARLARPQRRP
MSMEG_4127|M.smegmatis_MC2_155 AWILCAAIAHNLLRAAGVLAGTANAVARGSTLRRRIVTVPARLARPQRRP
Mvan_5550|M.vanbaalenii_PYR-1 AWILCAAIAYNLLRAVGVLAGGVHAVARGATLRRKFVNIPARLARPQRRA
Mflv_4617|M.gilvum_PYR-GCK -------------------------------------KPPAPLSVPRRLR
. ** *: *:*
MAV_1481|M.avium_104 ILHLPTHWPWADHWITLWCNTIGHSPPTTATI-
TH_0489|M.thermoresistible__bu VLHLPSHWPWADAWLRLWHNLFRERPPPQTVPA
MSMEG_4127|M.smegmatis_MC2_155 VLHLPTHWPWTDQWLMLWRNTIGYSPPATPCH-
Mvan_5550|M.vanbaalenii_PYR-1 ILHLPTHWPWAQQWLTLWRNTIGHSPPTAATH-
Mflv_4617|M.gilvum_PYR-GCK SRHG-----------------------------
*