For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIDVVTIFPAFLDALRQALPGKAIEAGIIDLAVHDLRDWTHDVHRSVDDSPYGGGPGMVMKAPVWGAALD EICSPETLLVVPSPAGRLFTQATAQRWSTERHLVFACGRYEGIDQRVVEDAATRMRVEEVSLGDYVLPGG ESAAVVMVEAVVRLLPDVLGNPASHQDDSHSEFVGGLLEGPSYTRPASWRGLDVPDVLLSGDHARIAAWR HQQGLERTRERRPDLLPPDAS*
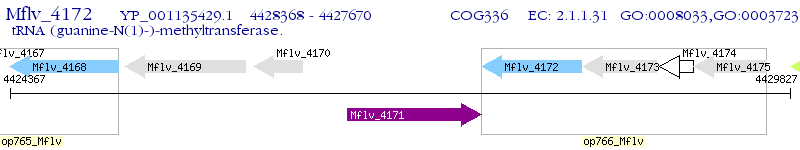
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4172 | trmD | - | 100% (232) | tRNA (guanine-N(1)-)-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2930c | trmD | 1e-113 | 84.35% (230) | tRNA (guanine-N(1)-)-methyltransferase |
| M. tuberculosis H37Rv | Rv2906c | trmD | 1e-113 | 84.35% (230) | tRNA (guanine-N(1)-)-methyltransferase |
| M. leprae Br4923 | MLBr_01615 | trmD | 1e-108 | 80.53% (226) | tRNA (guanine-N(1)-)-methyltransferase |
| M. abscessus ATCC 19977 | MAB_3226c | - | 2e-99 | 74.03% (231) | tRNA (guanine-N(1)-)-methyltransferase (TrmD) |
| M. marinum M | MMAR_1802 | trmD | 1e-107 | 79.13% (230) | tRNA (guanine-N1)-methyltransferase TrmD |
| M. avium 104 | MAV_3760 | trmD | 1e-108 | 81.94% (227) | tRNA (guanine-N(1)-)-methyltransferase |
| M. smegmatis MC2 155 | MSMEG_2438 | trmD | 1e-112 | 84.14% (227) | tRNA (guanine-N(1)-)-methyltransferase |
| M. thermoresistible (build 8) | TH_2387 | trmD | 1e-108 | 80.62% (227) | PROBABLE TRNA (GUANINE-N1)-METHYLTRANSFERASE TRMD |
| M. ulcerans Agy99 | MUL_2050 | trmD | 1e-108 | 79.57% (230) | tRNA (guanine-N(1)-)-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_2190 | trmD | 1e-125 | 93.39% (227) | tRNA (guanine-N(1)-)-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4172|M.gilvum_PYR-GCK MRIDVVTIFPAFLDALRQALPGKAIEAGIIDLAVHDLRDWTHDVHRSVDD
Mvan_2190|M.vanbaalenii_PYR-1 MRIDVVTIFPAFLDALRQALPGKAIEAGIVALAVHDLRDWTHDVHRSVDD
MMAR_1802|M.marinum_M MKIDVVTIFPAYLDPLRQSLPGRAIASGLVDLQVHDLRRWTYDVHHSVDD
MUL_2050|M.ulcerans_Agy99 MKIDVVTIFPAYLDPLRQSLPGRAIASGLVDLQVHDLRRWTYDVHHSVDD
MAV_3760|M.avium_104 MRIDVVTIFPAYLDPLRQSLPGKAIQSGLVDLRVHDLRRWTHDVHRSVDD
Mb2930c|M.bovis_AF2122/97 MRIDIVTIFPACLDPLRQSLPGKAIESGLVDLNVHDLRRWTHDVHHSVDD
Rv2906c|M.tuberculosis_H37Rv MRIDIVTIFPACLDPLRQSLPGKAIESGLVDLNVHDLRRWTHDVHHSVDD
MLBr_01615|M.leprae_Br4923 MKIDIVTIFPAYLDPLRQSLPGKAIESGLVDVQVHDLRRWTHDVHHSVDD
MSMEG_2438|M.smegmatis_MC2_155 MRIDVISIFPAYLDPIRQSLPGKAIDAGIVSVEVHDLRNWTHDVHRSVDD
TH_2387|M.thermoresistible__bu VRIDVITIFPRYLDPLRESLPGKAIEAGIVDLRVHDLRRWTHDVHRSVDD
MAB_3226c|M.abscessus_ATCC_199 MKIDVVTIFPEYLQPVRQSLPGKAIDAGLVDVAVHDLRRWTHDVHKSVDD
::**:::*** *:.:*::***:** :*:: : ***** **:***:****
Mflv_4172|M.gilvum_PYR-GCK SPYGGGPGMVMKAPVWGAALDEICSPETLLVVPSPAGRLFTQATAQRWST
Mvan_2190|M.vanbaalenii_PYR-1 SPYGGGPGMVMKAPIWGAALDEICSPETLLVVPSPAGRLFTQATAQRWSG
MMAR_1802|M.marinum_M SPYGGGPGMVMKAPVWGEALDEICSAETLLVVPTPAGSLFDQATARRWSA
MUL_2050|M.ulcerans_Agy99 SPYGGGPGMVMKAPVWGEALDEICSAETLLVVPTPAGSLFDQATAQRWSA
MAV_3760|M.avium_104 APYGGGPGMVMKAPVWGEALDEIASAETLLVVPTPAGALFDQATAARWSA
Mb2930c|M.bovis_AF2122/97 APYGGGPGMVMKAPVWGEALDEICSSETLLIVPTPAGVLFTQATAQRWTT
Rv2906c|M.tuberculosis_H37Rv APYGGGPGMVMKAPVWGEALDEICSSETLLIVPTPAGVLFTQATAQRWTT
MLBr_01615|M.leprae_Br4923 VPYGGGPGMVMKAPVWGEALDEICFDETLLVIPTPAGALFTQATAQCWST
MSMEG_2438|M.smegmatis_MC2_155 SPYGGGPGMVMKAPVWGEALDEICSEETLLVVPTPAGRLFDQRTAQRWST
TH_2387|M.thermoresistible__bu TPYGGGPGMVMTAPVWGEALDEICTEQTLLVVPTPAGRLFTQNVAQRWAG
MAB_3226c|M.abscessus_ATCC_199 SPYGGGPGMVMKPTVWGDALDEICTSETLLVVPTPAGYPFTQETAWQWST
**********...:** *****. :***::*:*** * * .* *:
Mflv_4172|M.gilvum_PYR-GCK ERHLVFACGRYEGIDQRVVEDAATRMRVEEVSLGDYVLPGGESAAVVMVE
Mvan_2190|M.vanbaalenii_PYR-1 EEHLVFACGRYEGIDQRVIDDAARRMRVEEVSIGDYVLPGGESAAVVMIE
MMAR_1802|M.marinum_M EQHLVFACGRYEGIDQRVAEDSARRMRVEEVSIGDYVLPGGESAAVVMIE
MUL_2050|M.ulcerans_Agy99 EQHLVFACGRYEGIDQRVAEDSARRMRVEEVSIGDYVLPGGESAAVVMIE
MAV_3760|M.avium_104 ERHLVFACGRYEGIDQRVVEDAARRMRVEEVSIGDYVLPGGESAAVVMIE
Mb2930c|M.bovis_AF2122/97 ESHLVFACGRYEGIDQRVVQDAARRMRVEEVSIGDYVLPGGESAAVVMVE
Rv2906c|M.tuberculosis_H37Rv ESHLVFACGRYEGIDQRVVQDAARRMRVEEVSIGDYVLPGGESAAVVMVE
MLBr_01615|M.leprae_Br4923 ERHLVFACGRYEGIDQRVVDDAVRRMRVEEVSIGDYVLPGGESAAVVMIE
MSMEG_2438|M.smegmatis_MC2_155 ERHLVFACGRYEGIDQRVVDDAARRMRVEEVSIGDYVLNGGEAATLVMVE
TH_2387|M.thermoresistible__bu ESHLVFACGRYEGIDQRVIDDAARRMRVEEVSIGDYVLAGGEAAALVMIE
MAB_3226c|M.abscessus_ATCC_199 EDHLVIACGRYEGIDQRVADDAATRMRVREVSIGDYVLNGGEAAALVIIE
* ***:************ :*:. ****.***:***** ***:*::*::*
Mflv_4172|M.gilvum_PYR-GCK AVVRLLPDVLGNPASHQDDSHSEFVGGLLEGPSYTRPASWRGLDVPDVLL
Mvan_2190|M.vanbaalenii_PYR-1 AVVRLLPDVLGNPASHQDDSHSELVGGLLEGPSYTRPASWRGLDVPDVLL
MMAR_1802|M.marinum_M AVLRLLTGVLGNPASHQEDSFSPELDRLLEGPSYTRPPSWRGLDVPEILL
MUL_2050|M.ulcerans_Agy99 AVLRLLTGVLGNPASHQEDSFSPELDRLLEGPSYTRPPSWRGLDVPEILL
MAV_3760|M.avium_104 AVLRLLDGVLGNPASRHDDSHSPALDRRLEGPSYTRPPSWRGLDVPEVLL
Mb2930c|M.bovis_AF2122/97 AVLRLLAGVLGNPASHQDDSHSTGLDGLLEGPSYTRPASWRGLDVPEVLL
Rv2906c|M.tuberculosis_H37Rv AVLRLLAGVLGNPASHQDDSHSTGLDGLLEGPSYTRPASWRGLDVPEVLL
MLBr_01615|M.leprae_Br4923 AVLRLVAGVLGNPASHRDDSHSPDLGRLLEGPSYTRPPTWRGLDVPPVLL
MSMEG_2438|M.smegmatis_MC2_155 AVVRLLPDVLGNPASHQQDSHS---DGLLEGPSYTRPPSWRGLDVPPVLL
TH_2387|M.thermoresistible__bu AVVRLLPGVMGNPASHQEDSHSGRYRGLLEGPSYTRPPSWRGLDVPEVLL
MAB_3226c|M.abscessus_ATCC_199 AVLRLVPGVLGNALSAQEDSHSEGMASLLEGPSYTRPPSWRGMDVPPVLL
**:**: .*:**. * ::**.* *********.:***:*** :**
Mflv_4172|M.gilvum_PYR-GCK SGDHARIAAWRHQQGLERTRERRPDLLP----PDAS------
Mvan_2190|M.vanbaalenii_PYR-1 SGDHARIAAWRREQGLQRTRERRPDLLD--------------
MMAR_1802|M.marinum_M SGDHARIATWRREVSLRRTRERRPDLAP----PE--------
MUL_2050|M.ulcerans_Agy99 SGDHARIATWRREVSLRRTRERRPDLAP----PE--------
MAV_3760|M.avium_104 SGDHARIAAWRREVSLQRTRERRPELLADPVGPQDGPGL---
Mb2930c|M.bovis_AF2122/97 SGDHARIAAWRREVSLQRTRERRPDLSH----PD--------
Rv2906c|M.tuberculosis_H37Rv SGDHARIAAWRREVSLQRTRERRPDLSH----PD--------
MLBr_01615|M.leprae_Br4923 SGDHARIAAWRREASLRRTRKRRPDLSG----ARFVWGLGLS
MSMEG_2438|M.smegmatis_MC2_155 SGDHAKVAAWRHEQALQRTRERRPDLLD--------------
TH_2387|M.thermoresistible__bu SGNHAKIAAWRREQALKRTRERRPDLLDSEGDPPGEQA----
MAB_3226c|M.abscessus_ATCC_199 SGDHAKIAAWRAEQSRQRTIERRPDLLGFDSPTGEHGGDGLS
**:**::*:** : . .** :***:*