For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTQKAGAPLPQAKPDAGASPRKGPNWGRWISAFALLGFFGLFTAFSRTELDPRVANPNVEGRPRPVEFLF GWDGWLWVHQIGCVILLVVLVAIFVRGWRRDPGSPVMLMFLCTTLIVWQDPIMNWAPFAVYNPELLHWPE NWPLIMLSPTVEPFIVFGYVAFYFAPFFPAIWCLRKLQAKHGPTAFVSRHPLISLGALTLVIGFIFDAIL EVSLVRTGLYIYSQAIPFGTLFPGTTFQFPLIWESLAVTFVMVPAAVLCYRDDTGKAVAEKLAARAKLFP SKPVLGTFLVMFFIINIAYFAYGSWFAAIKASGLATAVACPWPYPEAKVYDPQGYFEKEGAQGPFSVGKW ATWQSGLPNGRPEVTPPPPGEGDCAPENADG*
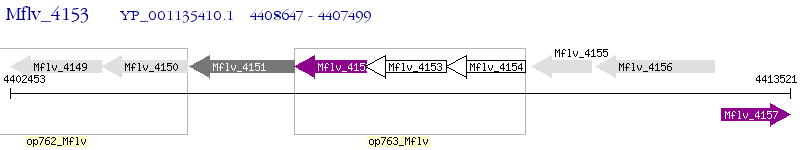
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4153 | - | - | 100% (382) | hypothetical protein Mflv_4153 |
| M. gilvum PYR-GCK | Mflv_2459 | - | 3e-11 | 28.36% (268) | hypothetical protein Mflv_2459 |
| M. gilvum PYR-GCK | Mflv_4146 | - | 4e-11 | 25.41% (244) | hypothetical protein Mflv_4146 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1272 | - | 1e-164 | 68.78% (378) | hypothetical protein MAV_1272 |
| M. smegmatis MC2 155 | MSMEG_6808 | - | 2e-11 | 28.73% (268) | hypothetical protein MSMEG_6808 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5888 | - | 1e-08 | 24.81% (258) | hypothetical protein Mvan_5888 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4153|M.gilvum_PYR-GCK --MTTQKAGAPLPQAKPDAGASPRKGPN-WGRWISAFALLGFFGLFTAFS
MAV_1272|M.avium_104 --MSTEQTTS--PAAQRESAAPRRTGTRGWGGWVAGAGLAAFALFFIANC
MSMEG_6808|M.smegmatis_MC2_155 MSELSDKKAAATESLRSVGRLDAPPAAKPSRRVKIWATAGGIILAFQIYV
Mvan_5888|M.vanbaalenii_PYR-1 ---MATNSRTTVSEDLTDPRAQKLVGTD-SPPVVWLARLGALFLVLELYI
: : : .. .: :
Mflv_4153|M.gilvum_PYR-GCK RTELDPRVANPNVEGRPRPVEFLFGWDGWLWVHQIGCVILLVVLVAIFVR
MAV_1272|M.avium_104 RVALDPRVANPNVQGRPRPVKFIFGLD-YMTFIQISTVVMLIVLVVVFVR
MSMEG_6808|M.smegmatis_MC2_155 WIRWITGPYFTRVDPGPTDPPTFMKIALMMWQIGS-PVLFPVAIWWFIIR
Mvan_5888|M.vanbaalenii_PYR-1 FGRWVTSDSFQRVMPGPDDGTN----MVWIWFWDVLSIGGSIVFVAWIVR
. .* * : : :.: ::*
Mflv_4153|M.gilvum_PYR-GCK GWRRDPGSPVMLMFLCTT-LIVWQDPIMN----WAPFAVYNPELLHWPEN
MAV_1272|M.avium_104 GWRRNPGSPAMLMFLCTT-LIVWQDPIMN----WAPFAVYNPDLIHWPES
MSMEG_6808|M.smegmatis_MC2_155 PWRRERRITLDGMIMVSTGLFFFQDPLLNYINTWCTYNAWAFNMGSWAPH
Mvan_5888|M.vanbaalenii_PYR-1 KRLREGELPPVAIVAMAWMLTAWQDPLVNALRPVFSYNSHFLNFGSWGEF
*: . :. : * :***::* .: :: *
Mflv_4153|M.gilvum_PYR-GCK WP--LIMLSP---TVEPF--IVFGYVAFYFAPFFPAIWCLRKLQAKHGPT
MAV_1272|M.avium_104 WP--LVSLSP---TVEPF--VVFGYVTFYFGPYFPAVWILRRLQAKYGPQ
MSMEG_6808|M.smegmatis_MC2_155 VPGWMSPEKPGAQVAEPLGINVSGYAYGVLLCTILGCWIMRKAQQRWPN-
Mvan_5888|M.vanbaalenii_PYR-1 IPGWIDNGGVN---PQPIFWWIGIYLFFVPLNMMGVDAYLNWVRRHLPR-
* : :*: : * : :. : :
Mflv_4153|M.gilvum_PYR-GCK AFVSRHPLISLGALTLVIGFIFDAILE-VSLVRTGLYIYSQAIPFGTLFP
MAV_1272|M.avium_104 SFFARHPLVSLGVLTCAIGFVFDAWLE-IQLVHMGMYIYSQVIPWGSVFT
MSMEG_6808|M.smegmatis_MC2_155 --LGTKGLIG---VAFAWGFVFDFVMEGLVLMPLGMYTFPGAIQSLSINA
Mvan_5888|M.vanbaalenii_PYR-1 --INRAGLIG---FLMILMFAQDVIGEWITLIQG-VDRYLWVGGSISLWP
. *:. . * * * : *: : : . :: .
Mflv_4153|M.gilvum_PYR-GCK GTTFQFPLIWESLAVT-FVMVPAAVLCYRDDTGKAVAEKLAARAKLFPSK
MAV_1272|M.avium_104 GTTFQFPLIWESFSVT-FVMVPAAILCYRDDTGKSVAEKLAAKAKLFPAR
MSMEG_6808|M.smegmatis_MC2_155 GTYYQWPIYEGLMWGG-VQAALCCLRFFTDDRGRTVVEKGLDNVRGGFAK
Mvan_5888|M.vanbaalenii_PYR-1 GTDYQFPLYEGIFWGCGMVGLSAMIYYFRDARGHMFTDRGLARLKIRRGR
** :*:*: : . . : : * *: ..:: . : .:
Mflv_4153|M.gilvum_PYR-GCK PVLGTFLVMFFIINIAYFAYGSWFAAIKASGLATAVACPWPYPEAK-VYD
MAV_1272|M.avium_104 PVLGTFLVMFVIINVSYFAYGAWFAVIKATKAATSVACPWPYPEAK-VYD
MSMEG_6808|M.smegmatis_MC2_155 QQLVRFLAIFAAISASFFTF-----YIVPAQFMSTHADPWPEDVQKRSYF
Mvan_5888|M.vanbaalenii_PYR-1 -TLIRVLALGAVFNAAMIVFN--LGYVAVNQHADTTQPTVPS------YF
* .*.: :. : :.: : : . * *
Mflv_4153|M.gilvum_PYR-GCK PQGYFEK-EGAQGPFSVGKWATWQSGLPNGRPEVTPPPPGEGDCAPENAD
MAV_1272|M.avium_104 PQGFYEK-AGAQGPYSVGIWSTWMSGQPHGRPDVQPPPPGAGACATTAVG
MSMEG_6808|M.smegmatis_MC2_155 TSGICGDGTDRPCPHPDLPIPTKHSGYINVDGELVLP---EGVEMPAVVP
Mvan_5888|M.vanbaalenii_PYR-1 QNGMCGLGDHPPCERGWW--------------------------------
.*
Mflv_4153|M.gilvum_PYR-GCK G-
MAV_1272|M.avium_104 HG
MSMEG_6808|M.smegmatis_MC2_155 HE
Mvan_5888|M.vanbaalenii_PYR-1 --