For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTEQTTSPAAQRESAAPRRTGTRGWGGWVAGAGLAAFALFFIANCRVALDPRVANPNVQGRPRPVKFIF GLDYMTFIQISTVVMLIVLVVVFVRGWRRNPGSPAMLMFLCTTLIVWQDPIMNWAPFAVYNPDLIHWPES WPLVSLSPTVEPFVVFGYVTFYFGPYFPAVWILRRLQAKYGPQSFFARHPLVSLGVLTCAIGFVFDAWLE IQLVHMGMYIYSQVIPWGSVFTGTTFQFPLIWESFSVTFVMVPAAILCYRDDTGKSVAEKLAAKAKLFPA RPVLGTFLVMFVIINVSYFAYGAWFAVIKATKAATSVACPWPYPEAKVYDPQGFYEKAGAQGPYSVGIWS TWMSGQPHGRPDVQPPPPGAGACATTAVGHG
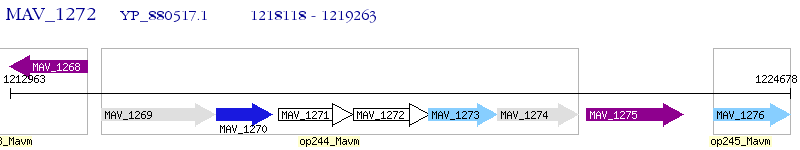
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1272 | - | - | 100% (381) | hypothetical protein MAV_1272 |
| M. avium 104 | MAV_2134 | - | 9e-12 | 27.85% (219) | hypothetical protein MAV_2134 |
| M. avium 104 | MAV_1951 | - | 3e-09 | 25.41% (244) | postpolyketide modification protein, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4153 | - | 1e-164 | 68.78% (378) | hypothetical protein Mflv_4153 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6808 | - | 6e-11 | 28.52% (277) | hypothetical protein MSMEG_6808 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4195 | - | 0.0001 | 30.36% (112) | hypothetical protein Mvan_4195 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6808|M.smegmatis_MC2_155 ---------------------MSELSDKKAAATESLRSVGRLDAPPAAKP
Mvan_4195|M.vanbaalenii_PYR-1 MQPAGSAAPPTASTIVKGKPIMSDLS-RKPAVTESVSGTADLGKQGPSQ-
MAV_1272|M.avium_104 -----------------------MSTEQTTS--PAAQRESAAPRRTGTRG
Mflv_4153|M.gilvum_PYR-GCK -----------------------MTTQKAGAPLPQAKPDAGASPRKGPN-
: : : . ..
MSMEG_6808|M.smegmatis_MC2_155 SRRVKIWATAGGIILAFQIYVWIRWITGPYFTRVDPGPTDPPTFMKIA-L
Mvan_4195|M.vanbaalenii_PYR-1 SNAFKVWATVGAVFLAYTLYVFIRWVSGPYFEPVAGGPSEPPLYMKIP-L
MAV_1272|M.avium_104 WGGWVAGAGLAAFALFFIANCRVALDPRVANPNVQGRPRPVKFIFGLD-Y
Mflv_4153|M.gilvum_PYR-GCK WGRWISAFALLGFFGLFTAFSRTELDPRVANPNVEGRPRPVEFLFGWDGW
.. : . * * :
MSMEG_6808|M.smegmatis_MC2_155 MMWQIGSPVLFPVAIWWFIIRPWRRERRITLDGMIMVSTGLFFFQDPLLN
Mvan_4195|M.vanbaalenii_PYR-1 MANAVVLWIGLPFALWFFLIRPWVREKRITLDGMLLVSMGLMMFQDPMLN
MAV_1272|M.avium_104 MTFIQISTVVMLIVLVVVFVRGWRRNPGSPAMLMFLCTT-LIVWQDPIMN
Mflv_4153|M.gilvum_PYR-GCK LWVHQIGCVILLVVLVAIFVRGWRRDPGSPVMLMFLCTT-LIVWQDPIMN
: : : ..: .::* * *: . *:: : *:.:***::*
MSMEG_6808|M.smegmatis_MC2_155 YINTWCTYNAWAFNMGSWAPHVPGWMSPEKPGAQVAEPLGINVSGYAYGV
Mvan_4195|M.vanbaalenii_PYR-1 YYSTWCTYNAWLFNQGSWAPHIPGWVAHEEPGHTVPEPLLTNIPGYMYGV
MAV_1272|M.avium_104 ----WAPFAVYNPDLIHWPESWP--LVSLSP--TVEPFVVFGYVTFYFGP
Mflv_4153|M.gilvum_PYR-GCK ----WAPFAVYNPELLHWPENWP--LIMLSP--TVEPFIVFGYVAFYFAP
*..: .: : *. * : .* * : . : :.
MSMEG_6808|M.smegmatis_MC2_155 LLCTILGCWIMRKAQQRWPNLGTKGLIGV---AFAWGFVFDFVMEGLVLM
Mvan_4195|M.vanbaalenii_PYR-1 LLLTIVGCALMRKIKNRWPGISNLRLVLV---TYAVAIVFDFIMEGLILL
MAV_1272|M.avium_104 YFPAVWILRRLQAKYGPQSFFARHPLVSLGVLTCAIGFVFDAWLE-IQLV
Mflv_4153|M.gilvum_PYR-GCK FFPAIWCLRKLQAKHGPTAFVSRHPLISLGALTLVIGFIFDAILE-VSLV
: :: :: . .. *: : : . .::** :* : *:
MSMEG_6808|M.smegmatis_MC2_155 PLGMYTFPGAIQSLSINAGTYYQWPIYEGLMWGGVQAALCCLRFFTDDRG
Mvan_4195|M.vanbaalenii_PYR-1 PIGFYSYPGAIQELSINAGTYYQYPIYEGFMWGGVQAALCCLRFFTDDRG
MAV_1272|M.avium_104 HMGMYIYSQVIPWGSVFTGTTFQFPLIWESFSVTFVMVPAAILCYRDDTG
Mflv_4153|M.gilvum_PYR-GCK RTGLYIYSQAIPFGTLFPGTTFQFPLIWESLAVTFVMVPAAVLCYRDDTG
*:* :. .* :: .** :*:*: : . . ..: : ** *
MSMEG_6808|M.smegmatis_MC2_155 RTVVEKGLDNVRGGFAKQQLVRFLAIFAAISASFFTF---YIVPAQFMST
Mvan_4195|M.vanbaalenii_PYR-1 RTVVERGLDRVRGGFVKQQFVRFLAIFGGVSACFFLF---YNVPATWLGM
MAV_1272|M.avium_104 KSVAEKLAAKAKLFPARPVLGTFLVMFVIINVSYFAYGAWFAVIKATKAA
Mflv_4153|M.gilvum_PYR-GCK KAVAEKLAARAKLFPSKPVLGTFLVMFFIINIAYFAYGSWFAAIKASGLA
::*.*: ..: : : **.:* :. .:* : : .
MSMEG_6808|M.smegmatis_MC2_155 HADPWPEDVQKRSYFTSGICGDGTDRPCPHPDLPIPTKHSGYINVDGELV
Mvan_4195|M.vanbaalenii_PYR-1 QGDPWPEDVQKRSYFNPGICGEGTDRPCPNPDLPLPTKHSGYVNHDGELV
MAV_1272|M.avium_104 TSVACPWPYPEAKVYDPQGFYEKAGAQGPYSVGIWSTWMSGQPHGRPDVQ
Mflv_4153|M.gilvum_PYR-GCK TAVACPWPYPEAKVYDPQGYFEKEGAQGPFSVGKWATWQSGLPNGRPEVT
. . * : . : . : . * . .* ** : ::
MSMEG_6808|M.smegmatis_MC2_155 LPEGVEMPAVVPHE--------
Mvan_4195|M.vanbaalenii_PYR-1 LPEGVEIPPVVPIQRPNSDDSQ
MAV_1272|M.avium_104 PPPPGAGACATTAVGHG-----
Mflv_4153|M.gilvum_PYR-GCK PPPPGEGDCAPENADG------
* .