For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TFGNYQLEIYLQGLSGIMPAYPMDFAGWEARAQASMPPSVWSYVAGGAGDENTQRANRTAFDRWGLMPRM LVGTTERDLTVDVFGLTLPSPIFMAPVGVAGICSQSGHGDLEAARAAARTGVPMAVSTLTEDPLEDVAAE FGDTPGFFQLYTPTDRDLAASFVQRAEAAGYKAIIVTLDTWIPGWRPRDLSTSNFPQLRGRCLSNYTSDP VFRGLLPQPPEENMQATVLQWAGMFGNALSWDDLPWLRSLTDLPLILKGLCHPDDVRRARDGGVDGIYCS THGGRQANGGLPAIDCLPVVVEAADGLPVLFDSGVRSGADIVKALALGATAVGIGRPYAYGLALGGTDGL VHVLRSLLAETDLIMAVDGYPTLADLTPDTVRRVT*
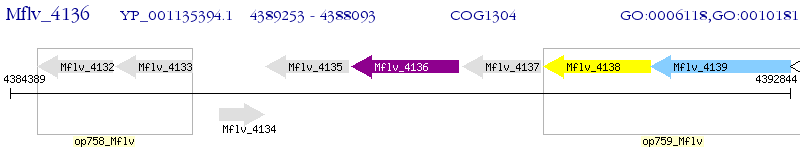
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4136 | - | - | 100% (386) | FMN-dependent alpha-hydroxy acid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5059 | - | 4e-45 | 32.44% (373) | (S)-2-hydroxy-acid oxidase |
| M. gilvum PYR-GCK | Mflv_3286 | - | 1e-05 | 32.04% (103) | 2-nitropropane dioxygenase, NPD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0713 | lldD1 | 7e-44 | 33.24% (370) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. tuberculosis H37Rv | Rv0694 | lldD1 | 8e-44 | 33.24% (370) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. leprae Br4923 | MLBr_02046 | lldD2 | 7e-42 | 32.88% (365) | L-lactate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2126 | - | 1e-101 | 51.05% (380) | putative L-lactate 2-monooxygenase |
| M. marinum M | MMAR_1816 | lldD1_1 | 0.0 | 80.52% (385) | L-lactate dehydrogenase (cytochrome) LldD1_1 |
| M. avium 104 | MAV_3747 | - | 0.0 | 80.78% (385) | lactate 2-monooxygenase |
| M. smegmatis MC2 155 | MSMEG_2512 | - | 0.0 | 81.56% (385) | lactate 2-monooxygenase |
| M. thermoresistible (build 8) | TH_0845 | - | 0.0 | 81.87% (386) | lactate 2-monooxygenase |
| M. ulcerans Agy99 | MUL_0774 | lldD1 | 1e-44 | 33.24% (376) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. vanbaalenii PYR-1 | Mvan_2206 | - | 0.0 | 87.01% (385) | FMN-dependent alpha-hydroxy acid dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4136|M.gilvum_PYR-GCK -----------MTFGNYQLEIYLQGLSGIMPAYPMDFAGWEARAQASMPP
Mvan_2206|M.vanbaalenii_PYR-1 -----------MTFGNYQLEIYLQGLSGILPNLPMDYAGWEAKAESAMPP
MMAR_1816|M.marinum_M -----------MAFGDYQYEIYFQGLSGVVPGLPMAFAELESRAQMALPP
MAV_3747|M.avium_104 -----------MAFGDYQNEIYFRGLGGVAPSLPMAFAELESRAERAMSP
TH_0845|M.thermoresistible__bu -----------MAFGDYQLEIYLQGLSGVVPSLPMAFAELEARARAAMSP
MSMEG_2512|M.smegmatis_MC2_155 -----------MAYGDYQLEIYLQGLAGVLPTMPMDYATLESKAQHAMPA
MAB_2126|M.abscessus_ATCC_1997 -----------MNFGDFQLQFYAAGALGKVSTLPFNFAELESRAEQTLGE
Mb0713|M.bovis_AF2122/97 -------------------------MAEAWFETVAIAQQR---AKRRLPK
Rv0694|M.tuberculosis_H37Rv -------------------------MAEAWFETVAIAQQR---AKRRLPK
MUL_0774|M.ulcerans_Agy99 -------------------------MADEWFETVAIAQQR---AKRRLPK
MLBr_02046|M.leprae_Br4923 MAAKRRMPRVHDLAPLIQFKRPKFGATKRRLEAALTIQDLRCIAKQRTPK
*.
Mflv_4136|M.gilvum_PYR-GCK SVWSYVAGGAGDENTQRANRTAFDRWGLMPRMLVGTTERDLTVDVFGLTL
Mvan_2206|M.vanbaalenii_PYR-1 SVWSYVVGGAGDEHTQRANRTAFDRWGLIPRMLVGATERDLTVDLFGMTL
MMAR_1816|M.marinum_M SVWSYVVGGAGDERTQRANCEAFDRWGLMPRMFVGAAERDLTVEMFGLTL
MAV_3747|M.avium_104 SVWSYVTGGAGDERTQRANREAFDRWGLIPRMFVGAAERDLSVQMFGLTL
TH_0845|M.thermoresistible__bu SVWSYVAGGAGDERTQRINVSAFDNWGLIPRMLVGATERDLSVNLFGLTL
MSMEG_2512|M.smegmatis_MC2_155 SIWSYVAGGAGDERTQRVNRTAFDRWGLVPRMLNAQRERDLSVDLFGLQL
MAB_2126|M.abscessus_ATCC_1997 GIFGYVRGGAGDEHTQDSNVTALRRYGLVPRMLRDRTVRDMSTSFLGRTF
Mb0713|M.bovis_AF2122/97 SVYSSLIAASEKGITVADNVAAFSELGFAPHVIGATDKRDLSTTVMGQEV
Rv0694|M.tuberculosis_H37Rv SVYSSLIAASEKGITVADNVAAFSELGFAPHVIGATDKRDLSTTVMGQEV
MUL_0774|M.ulcerans_Agy99 SVYSSLISASEKGITVADNVAAFSELGFAPHVIGAAEKRDMSTTVMGQDI
MLBr_02046|M.leprae_Br4923 AAFDYTDGAAEDELSIKRAQQAFRDIEFHPAILRDVTNVCAGWDVLGHSV
. :. ..: . : *: : * :: .:* .
Mflv_4136|M.gilvum_PYR-GCK PSPIFMAPVGVAGICSQSGHGDLEAARAAARTGVPMAVSTLTEDPLEDVA
Mvan_2206|M.vanbaalenii_PYR-1 PSPLFMAPIGVIGICSQNGQGDLAAARAAARTGVPMTVSTLTEDPLEDVA
MMAR_1816|M.marinum_M PSPIFLAPIGVIGLCAQDGHGDLATARAAARTGVPMVVSTLTADPMEDVA
MAV_3747|M.avium_104 PSPVFMAPIGVIGICAQDGHGDLATARAAAATGVPMVVSTLTADPMEDVA
TH_0845|M.thermoresistible__bu PSPIFMAPIGVLGICAQDHHGDLACARAAARTGVPLVVSTLTQDPLEDIA
MSMEG_2512|M.smegmatis_MC2_155 PSPLFMAPIGVLGICGQDGHGDLAGAQAAARTGVPMVVSTLTQDPLENVA
MAB_2126|M.abscessus_ATCC_1997 TSPAFICPVGVLGAVRD--RGDLLTAAAARELDMPAMYSTLSAATLEEVA
Mb0713|M.bovis_AF2122/97 SLPVIISPTG-VQAVDP--GGEVAVARAAAARGTVMGLSSFASKPIEEVI
Rv0694|M.tuberculosis_H37Rv SLPVIISPTG-VQAVDP--GGEVAVARAAAARGTVMGLSSFASKPIEEVI
MUL_0774|M.ulcerans_Agy99 SMPVLISPTG-VQAVHP--DGEVAVARAAAARGTAMGLSSFASKTIEDVI
MLBr_02046|M.leprae_Br4923 LLPFGIAPTGFTRLMHT--EGEIAGARAAAAAGIPFSLSTLATSAIEDVV
* :.* * *:: * ** . *::: .:*::
Mflv_4136|M.gilvum_PYR-GCK AEFGDTPGFFQLYTPTDRDLAASFVQRAEAAGYKAIIVTLDTWIPGWRPR
Mvan_2206|M.vanbaalenii_PYR-1 AEFGDTPGFFQLYTPTDRDLAASLVHRAEAAGYKAIIVTLDTWIPGWRPR
MMAR_1816|M.marinum_M AEFGDIPGFFQLYTPKDRELAASLVQRAESAGFKGIVVTLDTWIPGWRPR
MAV_3747|M.avium_104 AQFGDTPGFFQLYTPKDRELAASLVHRAEAAGFKGIIVTLDTWIPGWRPR
TH_0845|M.thermoresistible__bu AEFGDTPGFFQLYTPNDRDLAASLVRRAEKAGYKGIIVTLDTWIPGWRPR
MSMEG_2512|M.smegmatis_MC2_155 AQFGDTPGFFQLYTPTDRDLAASFVRRAEAAGYKAIVVTLDTWVPGWRPR
MAB_2126|M.abscessus_ATCC_1997 AARGDSYGIFQLYPSSDSELTDNFIRRAEAAGYDALAVTLDTGTLGWRPR
Mb0713|M.bovis_AF2122/97 AANP--KTFFQVYWQGGRDALAERVERARQAGAVGLVVTTDWTFSHGRDW
Rv0694|M.tuberculosis_H37Rv AANP--KTFFQVYWQGGRDALAERVERARQAGAVGLVVTTDWTFSHGRDW
MUL_0774|M.ulcerans_Agy99 AANP--KIFFQIYWLGGRDAIAERVERARQAGAVGLIVTTDWTFSHGRDW
MLBr_02046|M.leprae_Br4923 AAVPQGRKWFQLYMWRDRDRSMALVERAADAGYDALLVTVDVPVAGARLR
* **:* . : :.** ** .: ** * *
Mflv_4136|M.gilvum_PYR-GCK DLSTS-------NFPQLRGRCLSNYTSDPVFR--GLLPQPPEENMQA---
Mvan_2206|M.vanbaalenii_PYR-1 DLAMS-------NFPQLRGRCLANYTSDPVFR--AALSQPPEENMQA---
MMAR_1816|M.marinum_M DLSTA-------NFPQLRGHCLSNYTSDPVFR--AGLPRPPEEDPQG---
MAV_3747|M.avium_104 DLSTA-------NFPQLRGMCLSNYTSDPVFR--AALARPPEEDPQG---
TH_0845|M.thermoresistible__bu DLSTS-------NFPQLRGYSLANYTSDPVFS--AGLQQPPEQNPQA---
MSMEG_2512|M.smegmatis_MC2_155 DLSTS-------NFPQLRGKCLANYTSDPVFR--AGLPQPPEENPQA---
MAB_2126|M.abscessus_ATCC_1997 DLKHG-------YLPMLHGHCLANYTSDPRFLEIAGVRSAGELTPMH---
Mb0713|M.bovis_AF2122/97 GSPK---IPEEMNLKTILRLSPEAITRPRWLWKFAKTLRPPDLRVPNQGR
Rv0694|M.tuberculosis_H37Rv GSPK---IPEEMNLKTILRLSPEAITRPRWLWKFAKTLRPPDLRVPNQGR
MUL_0774|M.ulcerans_Agy99 GSPK---IPEQMNLRTILRLSPEAIVRPRWLWKFGKTLRPPDLRVPNQGR
MLBr_02046|M.leprae_Br4923 DTRNGMSIPPALTLRTVF----DALPHPRWWIDLLTTEP---LAFASLDR
. : :
Mflv_4136|M.gilvum_PYR-GCK -------TVLQWAGMFGNA-LSWDDLPWLRSLTDLPLILKGLCHPDDVRR
Mvan_2206|M.vanbaalenii_PYR-1 -------AVLQWVAQFGNA-LTWEDLPWLRSLTDLPLVLKGLCHPDDVRR
MMAR_1816|M.marinum_M -------TVLRWAQLFGNP-LTWSDLAWLRELTDLPLIVKGICHPDDARR
MAV_3747|M.avium_104 -------TVLQWISTFGNP-LTWDDLPWLRSLTDLPLIIKGICHPDDARR
TH_0845|M.thermoresistible__bu -------VVTRWVQLFGNS-LTWDDLPWLRSLTTLPLILKGICHPDDVRR
MSMEG_2512|M.smegmatis_MC2_155 -------TVLRWVSQFGNP-LTWADLPWLRSLTKLPLILKGICHPDDVRR
MAB_2126|M.abscessus_ATCC_1997 -------AGLVWASLFSHPGLTWADIDHYRSITKLPIILKGICDSDDVRQ
Mb0713|M.bovis_AF2122/97 RGEPGPPFFAAYGEWMATPPPTWEDIGWLRELWGGPFMLKGVMRVDDAKR
Rv0694|M.tuberculosis_H37Rv RGEPGPPFFAAYGEWMATPPPTWEDIGWLRELWGGPFMLKGVMRVDDAKR
MUL_0774|M.ulcerans_Agy99 RGEPGPAFFAAYGEWMGTPPPTWDDIAWLRELWGGPFMLKGVMRVDDAKR
MLBr_02046|M.leprae_Br4923 WSG----TVSDYLNTMFDPSVTFDDLAWIKTQWPGKFVVKGIQTLDDARA
: : . :: *: : :::**: **.:
Mflv_4136|M.gilvum_PYR-GCK ARDGGVDGIYCSTHGGRQANGGLPAIDCLPVVVEAADG-LPVLFDSGVRS
Mvan_2206|M.vanbaalenii_PYR-1 AKDAGVDGVYCSTHGGRQANGGLPALDCLPGVVEAADG-LPVLFDSGIRS
MMAR_1816|M.marinum_M AKDGGVDGIYCSTHGGRQANGGLPALDCLPGVVEAADG-LPVLFDSGIRS
MAV_3747|M.avium_104 ARDGGVDGIYCSTHGGRQANGGLPALDCLPGVVEAADG-LPVLFDSGIRS
TH_0845|M.thermoresistible__bu AKDAGVDGIYCSNHGGRQANGGLPAIDCLPDVVAAADG-MPVLFDSGIRS
MSMEG_2512|M.smegmatis_MC2_155 AKDEGVDGIYCSNHGGRQANGGVPAIDCLPGVVEAADG-LPVLFDSGIRN
MAB_2126|M.abscessus_ATCC_1997 AVDRGVDAIAYSNHGGRQANGGVPAIDGLAAAVEAAGS-VPVTFDSGIRD
Mb0713|M.bovis_AF2122/97 AVDAGVSAISVSNHGGNNLDGTPASIRALPAVSAAVGDQVEVLLDGGIRR
Rv0694|M.tuberculosis_H37Rv AVDAGVSAISVSNHGGNNLDGTPASIRALPAVSAAVGDQVEVLLDGGIRR
MUL_0774|M.ulcerans_Agy99 AVDAGVSAISVSNHGGNNLDGTPASIRALPAVAAAVGDQVEVLLDGGIRR
MLBr_02046|M.leprae_Br4923 VVERGIDGVVLSNHGGRQLDRAPVPFHLLPTVAREFGKDTEILLDTGIMS
. : *:..: *.***.: : .: *. . . : :* *:
Mflv_4136|M.gilvum_PYR-GCK GADIVKALALGATAVGIGRPYAYGLALGGTDGLVHVLRSLLAETDLIMAV
Mvan_2206|M.vanbaalenii_PYR-1 GADIVKALALGATAVGVGRPYAYGLAIGGEDGIVHVLRSLLAEADLIMAV
MMAR_1816|M.marinum_M GADIIKALAMGATAVGIGRPYAYGLALGGVDGVVHVLRMLLAEADLIMAV
MAV_3747|M.avium_104 GADVVKALALGATAVGIGRPYAYGLALGGVDGIVHVLRSILAEADLIMAV
TH_0845|M.thermoresistible__bu GADIVKALALGATAVGIGRPYPYGLALGGEDGVVHVLRSLLAEADLIMAV
MSMEG_2512|M.smegmatis_MC2_155 GADIVKALALGATAVGVGRPYVFGLALGGVDGIVHVLRSLLAEADLIMAV
MAB_2126|M.abscessus_ATCC_1997 GIDILRAVALGASLVGVARPYVYGLALDGTNGVKHVIQSLLAEADLTMAV
Mb0713|M.bovis_AF2122/97 GSDVVKAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMG
Rv0694|M.tuberculosis_H37Rv GSDVVKAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMG
MUL_0774|M.ulcerans_Agy99 GSDVVKAVALGARAVLVGRAYLWGLAANGQAGVENVLDILRGGIDSALMG
MLBr_02046|M.leprae_Br4923 GADIVAAIALGARCTLVGRAYLYGLMAGGEAGVRRAIEILESGVIRTMQL
* *:: *:*:** . :.*.* :** .* *: ..: : . :
Mflv_4136|M.gilvum_PYR-GCK DGYPTLADLTPDTVRRVT--------------
Mvan_2206|M.vanbaalenii_PYR-1 DGYRSLADLTPDTLRRVI--------------
MMAR_1816|M.marinum_M DGYPTRKDLTPDTLRRVN--------------
MAV_3747|M.avium_104 DGYPTRKDLTPDTLRRVV--------------
TH_0845|M.thermoresistible__bu DGYPTLADLTPDALRRVT--------------
MSMEG_2512|M.smegmatis_MC2_155 DGYPSLADLTPDTLRRVDA-------------
MAB_2126|M.abscessus_ATCC_1997 NCYLSLNELAVQRLS-----------------
Mb0713|M.bovis_AF2122/97 LGHASVHDLSPADILVPTGFIRDLGVPSRRDV
Rv0694|M.tuberculosis_H37Rv LGHASVHDLSPADILVPTGFIRDLGVPSRRDV
MUL_0774|M.ulcerans_Agy99 LGHSSIHDLRSDDILIPADFVRRLGR------
MLBr_02046|M.leprae_Br4923 LGVTCLEELSPR----HVTQLQRLGPIEPPA-
:*