For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAEAWFETVAIAQQRAKRRLPKSVYSSLIAASEKGITVADNVAAFSELGFAPHVIGATDKRDLSTTVMGQ EVSLPVIISPTGVQAVDPGGEVAVARAAAARGTVMGLSSFASKPIEEVIAANPKTFFQVYWQGGRDALAE RVERARQAGAVGLVVTTDWTFSHGRDWGSPKIPEEMNLKTILRLSPEAITRPRWLWKFAKTLRPPDLRVP NQGRRGEPGPPFFAAYGEWMATPPPTWEDIGWLRELWGGPFMLKGVMRVDDAKRAVDAGVSAISVSNHGG NNLDGTPASIRALPAVSAAVGDQVEVLLDGGIRRGSDVVKAVALGARAVMIGRAYLWGLAANGQAGVENV LDILRGGIDSALMGLGHASVHDLSPADILVPTGFIRDLGVPSRRDV
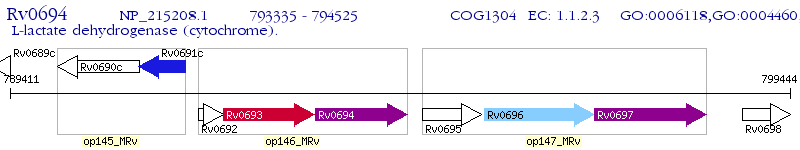
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0694 | lldD1 | - | 100% (396) | POSSIBLE L-LACTATE DEHYDROGENASE (CYTOCHROME) LLDD1 |
| M. tuberculosis H37Rv | Rv1872c | lldD2 | 4e-53 | 35.06% (385) | L-lactate dehydrogenase (cytochrome) LldD2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0713 | lldD1 | 0.0 | 100.00% (396) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. gilvum PYR-GCK | Mflv_5059 | - | 0.0 | 81.19% (388) | (S)-2-hydroxy-acid oxidase |
| M. leprae Br4923 | MLBr_02046 | lldD2 | 2e-56 | 34.30% (379) | L-lactate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3834c | - | 0.0 | 77.40% (385) | L-lactate dehydrogenase LldD1 |
| M. marinum M | MMAR_1022 | lldD1 | 0.0 | 89.20% (389) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. avium 104 | MAV_4477 | - | 0.0 | 88.75% (391) | FMN-dependent dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1424 | - | 0.0 | 81.49% (389) | FMN-dependent dehydrogenase |
| M. thermoresistible (build 8) | TH_2565 | - | 0.0 | 82.40% (392) | PUTATIVE FMN-dependent dehydrogenase |
| M. ulcerans Agy99 | MUL_0774 | lldD1 | 0.0 | 89.20% (389) | L-lactate dehydrogenase (cytochrome) LldD1 |
| M. vanbaalenii PYR-1 | Mvan_1297 | - | 0.0 | 79.59% (387) | (S)-2-hydroxy-acid oxidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5059|M.gilvum_PYR-GCK ------------------------MARDTWFETVAIAQQR---AKKRLPK
Mvan_1297|M.vanbaalenii_PYR-1 ------------------------MARDTWFETVAIAQQR---AKKRLPK
MSMEG_1424|M.smegmatis_MC2_155 ----------------------MNMARDIWFETVAIAQQR---ARKRLPK
Rv0694|M.tuberculosis_H37Rv ------------------------MA-EAWFETVAIAQQR---AKRRLPK
Mb0713|M.bovis_AF2122/97 ------------------------MA-EAWFETVAIAQQR---AKRRLPK
MMAR_1022|M.marinum_M ------------------------MA-DEWFETVAIAQQR---AKRRLPK
MUL_0774|M.ulcerans_Agy99 ------------------------MA-DEWFETVAIAQQR---AKRRLPK
MAV_4477|M.avium_104 ------------------------MA-DEWFETVAIAQQR---AKRRLPK
TH_2565|M.thermoresistible__bu ----------------------VTMARDKWFETVSIAQER---AKKRLPK
MAB_3834c|M.abscessus_ATCC_199 ------------------------MARNKWFETVAIAQQR---AKKRLPK
MLBr_02046|M.leprae_Br4923 MAAKRRMPRVHDLAPLIQFKRPKFGATKRRLEAALTIQDLRCIAKQRTPK
* . :*:. *: *::* **
Mflv_5059|M.gilvum_PYR-GCK SAYSSLISASEKGVSVSDNVEAFAELGFAPHVVGATDKRDMATTVMGQDI
Mvan_1297|M.vanbaalenii_PYR-1 SAYSSLISASEKGVTVTDNVESFAELGFAPHVIGATEKRDMATSVLGQDI
MSMEG_1424|M.smegmatis_MC2_155 SVYSSLISASEKGVTVTDNVESFAELGFAPHVVGAPEKRDMATTVMGQQI
Rv0694|M.tuberculosis_H37Rv SVYSSLIAASEKGITVADNVAAFSELGFAPHVIGATDKRDLSTTVMGQEV
Mb0713|M.bovis_AF2122/97 SVYSSLIAASEKGITVADNVAAFSELGFAPHVIGATDKRDLSTTVMGQEV
MMAR_1022|M.marinum_M SVYSSLISASEKGITVADNVAAFSELGFAPHVIGAAEKRDMSTTVMGQDI
MUL_0774|M.ulcerans_Agy99 SVYSSLISASEKGITVADNVAAFSELGFAPHVIGAAEKRDMSTTVMGQDI
MAV_4477|M.avium_104 SVYAALIAASEKGITVSDNVEAFGELGFAPHVVGAPAKRELATTVMGQEI
TH_2565|M.thermoresistible__bu SVYGALIAGSEKGLTVSDNVEAFGELGFAPHVVGGPAKRDMATTVMGQEI
MAB_3834c|M.abscessus_ATCC_199 SVYSSLISASEKGLTVSDNVEAFGELGFEPHVVGIQPDRELSTTVLGQDI
MLBr_02046|M.leprae_Br4923 AAFDYTDGAAEDELSIKRAQQAFRDIEFHPAILRDVTNVCAGWDVLGHSV
:.: ..:*. ::: :* :: * * :: . . *:*:.:
Mflv_5059|M.gilvum_PYR-GCK PLPVIISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKPMEEVTAV
Mvan_1297|M.vanbaalenii_PYR-1 SLPVIISPTG-VQAIDPDGEVAVARAAAARGTAMGLSSFASKPMEDVTAV
MSMEG_1424|M.smegmatis_MC2_155 SLPVIISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKPIEEVVAV
Rv0694|M.tuberculosis_H37Rv SLPVIISPTG-VQAVDPGGEVAVARAAAARGTVMGLSSFASKPIEEVIAA
Mb0713|M.bovis_AF2122/97 SLPVIISPTG-VQAVDPGGEVAVARAAAARGTVMGLSSFASKPIEEVIAA
MMAR_1022|M.marinum_M SMPVLISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKTIEDVIAA
MUL_0774|M.ulcerans_Agy99 SMPVLISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKTIEDVIAA
MAV_4477|M.avium_104 SMPVLISPTG-VQAVDPDGEVAVARAAAARGTAMGLSSFASKPIEEVIAV
TH_2565|M.thermoresistible__bu SLPVVISPTG-VQAVHPDGEVAVARAAAARGTAMGLSSFASKPIEEVIAA
MAB_3834c|M.abscessus_ATCC_199 SLPVMISPTG-VQAVDPDGEVAVARAAAARGTAMGLSSFASKPIEDVVAA
MLBr_02046|M.leprae_Br4923 LLPFGIAPTGFTRLMHTEGEIAGARAAAAAGIPFSLSTLATSAIEDVVAA
:*. *:*** .: :.. **:* ****** * :.**::*:..:*:* *.
Mflv_5059|M.gilvum_PYR-GCK ND--KIFFQIYWLGSRDAIAARMERARAAGAKGLILTTDWSFSHGRDWGS
Mvan_1297|M.vanbaalenii_PYR-1 ND--KIFFQIYWLGSRDDILARMERARAAGAKGLILTTDWSFAHGRDWGS
MSMEG_1424|M.smegmatis_MC2_155 ND--KIFFQIYWLGDRDAILARAERAKAAGAVGLIVTTDWSFSHGRDWGS
Rv0694|M.tuberculosis_H37Rv NP--KTFFQVYWQGGRDALAERVERARQAGAVGLVVTTDWTFSHGRDWGS
Mb0713|M.bovis_AF2122/97 NP--KTFFQVYWQGGRDALAERVERARQAGAVGLVVTTDWTFSHGRDWGS
MMAR_1022|M.marinum_M NP--KIFFQIYWLGGRDAIAERVERARQAGAVGLIVTTDWTFSHGRDWGS
MUL_0774|M.ulcerans_Agy99 NP--KIFFQIYWLGGRDAIAERVERARQAGAVGLIVTTDWTFSHGRDWGS
MAV_4477|M.avium_104 NP--KVFFQVYWLGGRDAIAERVERARQAGAVGLIVTTDWSFSHGRDWGS
TH_2565|M.thermoresistible__bu NP--KLFFQVYWLGGRDAIAARVERAREAGAAGLIVTTDWSFSHGRDWGS
MAB_3834c|M.abscessus_ATCC_199 NP--KTHFQIYWLGGRDDVAQRIQRAKDAGAVGLIATLDWSFSHGRDWGS
MLBr_02046|M.leprae_Br4923 VPQGRKWFQLYMWRDRDRSMALVERAADAGYDALLVTVDVPVAGARLR-D
: **:* .** :** ** .*: * * ..: .* .
Mflv_5059|M.gilvum_PYR-GCK PKIPEQMDLKTILKMSPEVLTKPRWFFSFAKTLRPPDLRVPNQGRPGEPG
Mvan_1297|M.vanbaalenii_PYR-1 PKIPERMDLKTMIRMSPEVITKPRWFFSFAKHLRPPDLRVPNQGRRGEAG
MSMEG_1424|M.smegmatis_MC2_155 PKIPEKMDLKTMVTMMPEALTKPRWLWQWGKTMRPPNLRVPNQGARGEDG
Rv0694|M.tuberculosis_H37Rv PKIPEEMNLKTILRLSPEAITRPRWLWKFAKTLRPPDLRVPNQGRRGEPG
Mb0713|M.bovis_AF2122/97 PKIPEEMNLKTILRLSPEAITRPRWLWKFAKTLRPPDLRVPNQGRRGEPG
MMAR_1022|M.marinum_M PKIPEQMNLRTILRLSPEAIVRPRWLWKFGKTLRPPDLRVPNQGRRGEPG
MUL_0774|M.ulcerans_Agy99 PKIPEQMNLRTILRLSPEAIVRPRWLWKFGKTLRPPDLRVPNQGRRGEPG
MAV_4477|M.avium_104 PKIPEEMNLRTILRLSPEAVFKPRWLWKFGKTLRPPELRVPNQGRRGEPG
TH_2565|M.thermoresistible__bu PKIPESMNLRTILRMLPEGLSRPRWMMQWAKTLRPPNLRVPNQAARGEAG
MAB_3834c|M.abscessus_ATCC_199 PAIPEKMDLRSMIRLAPEVVTKPSWLWSFGKGMNIPDLRVPNQAARGEAG
MLBr_02046|M.leprae_Br4923 TRNGMSIPPALTLRTVFDALPHPRWWIDLLTTEPLAFASLDRWSG-----
. : : : : :* * . . . : . .
Mflv_5059|M.gilvum_PYR-GCK PTFFEAYGEWMGTPPPTWEDVAWLREQWGGPFLLKGLVRVDDAKRAVDAG
Mvan_1297|M.vanbaalenii_PYR-1 PTFFEAYGQWMGTPPPTWEDVAWLREQWGGPFLLKGTVRVDDAKRAVDAG
MSMEG_1424|M.smegmatis_MC2_155 PPFFQAYGEWMGTPPPTWEDIAWLREQWDGPFMLKGVIRVDDAKRAVDAG
Rv0694|M.tuberculosis_H37Rv PPFFAAYGEWMATPPPTWEDIGWLRELWGGPFMLKGVMRVDDAKRAVDAG
Mb0713|M.bovis_AF2122/97 PPFFAAYGEWMATPPPTWEDIGWLRELWGGPFMLKGVMRVDDAKRAVDAG
MMAR_1022|M.marinum_M PAFFAAYGEWMGTPPPTWDDIAWLRELWGGPFMLKGVMRVDDAKRAVDAG
MUL_0774|M.ulcerans_Agy99 PAFFAAYGEWMGTPPPTWDDIAWLRELWGGPFMLKGVMRVDDAKRAVDAG
MAV_4477|M.avium_104 PPFFAAYGEWMGTPPPTWEDIAWLREVWGGPFMLKGVMRVDDAKRAVDAG
TH_2565|M.thermoresistible__bu PPFFHAYGEWMGTPPPTWDDIAWLRELWGGPFMLKGVIRVDDAKRAVDAG
MAB_3834c|M.abscessus_ATCC_199 PPFFDAYGQWMGTPAPTWDDVRWMREQWDGPFMLKGVMRIDDAKRAVDCG
MLBr_02046|M.leprae_Br4923 --TVSDYLNTMFDPSVTFDDLAWIKTQWPGKFVVKGIQTLDDARAVVERG
. * : * *. *::*: *:: * * *::** :***: .*: *
Mflv_5059|M.gilvum_PYR-GCK VSAITVSNHGGNNLDGTPAAIRCLPAIADAVGDQVEVLLDGGIRRGSDVV
Mvan_1297|M.vanbaalenii_PYR-1 VSAITVSNHGGNNLDGTPAAIRCLPAIADAVGDQVEVLLDGGIRRGSDVV
MSMEG_1424|M.smegmatis_MC2_155 VSAISVSNHGGNNLDGTPAAIRALPVIAEAVGDQVEVLLDGGIRRGSDVV
Rv0694|M.tuberculosis_H37Rv VSAISVSNHGGNNLDGTPASIRALPAVSAAVGDQVEVLLDGGIRRGSDVV
Mb0713|M.bovis_AF2122/97 VSAISVSNHGGNNLDGTPASIRALPAVSAAVGDQVEVLLDGGIRRGSDVV
MMAR_1022|M.marinum_M VSAISVSNHGGNNLDGTPASIRALPAVAAAVGDQVEVLLDGGIRRGSDVV
MUL_0774|M.ulcerans_Agy99 VSAISVSNHGGNNLDGTPASIRALPAVAAAVGDQVEVLLDGGIRRGSDVV
MAV_4477|M.avium_104 VSAISVSNHGGNNLDGTPASIRALPAIAEAVGDQIEVLLDGGVRRGSDVV
TH_2565|M.thermoresistible__bu VSAISVSNHGGNNLDSTPAAIRALPAIVDAVGDQVEVLLDGGIRRGSDVV
MAB_3834c|M.abscessus_ATCC_199 VSAISVSNHGGNNLDGTPASIRALPGIADAVGHDIEVLLDGGIRRGSDVV
MLBr_02046|M.leprae_Br4923 IDGVVLSNHGGRQLDRAPVPFHLLPTVAREFGKDTEILLDTGIMSGADIV
:..: :*****.:** :*..:: ** : .*.: *:*** *: *:*:*
Mflv_5059|M.gilvum_PYR-GCK KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILSGGIDSALRGLGKSS
Mvan_1297|M.vanbaalenii_PYR-1 KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGKSS
MSMEG_1424|M.smegmatis_MC2_155 KAVALGARAVMIGRAYLWGLAAEGQVGVENVLDILRGGIDSALMGLGRSS
Rv0694|M.tuberculosis_H37Rv KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGHAS
Mb0713|M.bovis_AF2122/97 KAVALGARAVMIGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGHAS
MMAR_1022|M.marinum_M KAVALGARAVLVGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGHSS
MUL_0774|M.ulcerans_Agy99 KAVALGARAVLVGRAYLWGLAANGQAGVENVLDILRGGIDSALMGLGHSS
MAV_4477|M.avium_104 KAVALGARAVMIGRAYLWGLAAAGQAGVENVLDILRGGIDSALMGLGHSS
TH_2565|M.thermoresistible__bu KAVALGARAVMIGRAYLWGLAAEGQAGVENVLDILRGGIDSALLGLGRAS
MAB_3834c|M.abscessus_ATCC_199 KALALGARAVMIGRAYLWGLAASGQAGVENVLDIMRGGIDSALMGLGKKS
MLBr_02046|M.leprae_Br4923 AAIALGARCTLVGRAYLYGLMAGGEAGVRRAIEILESGVIRTMQLLGVTC
*:*****..::*****:** * *:.**...::*: .*: :: ** .
Mflv_5059|M.gilvum_PYR-GCK IQELTPEDILVPEGFTRTLGV------
Mvan_1297|M.vanbaalenii_PYR-1 IHELTREDILIPDGFTRTLGA------
MSMEG_1424|M.smegmatis_MC2_155 IHDLVPEDILVPEGFTRALGVPPASGS
Rv0694|M.tuberculosis_H37Rv VHDLSPADILVPTGFIRDLGVPSRRDV
Mb0713|M.bovis_AF2122/97 VHDLSPADILVPTGFIRDLGVPSRRDV
MMAR_1022|M.marinum_M IHDLRSDDILIPADFVRRLGG------
MUL_0774|M.ulcerans_Agy99 IHDLRSDDILIPADFVRRLGR------
MAV_4477|M.avium_104 VHDLGPSDILVPPGFTRALGVPPAG--
TH_2565|M.thermoresistible__bu VHDLTPDDVVVPPGFTRALGVDPTRP-
MAB_3834c|M.abscessus_ATCC_199 VHELSPDDLLIPEGFARGLGRH-----
MLBr_02046|M.leprae_Br4923 LEELSPRHVTQLQRLGPIEPPA-----
:.:* .: :