For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PKLSAGLLLFRETDGVVEVLLGHPGGPFWARKDEGAWSIPKGEYADGDDPWAVAQREFTEETGKPVPAGP PIGLAPVRQPGGKVVTAFAVRGDLDLDGTFSNTFTLVWPRGSGTVREFPEIDRVEWFDLATARVKLLKGQ RPLLDELEKVLAHDGPDASRSR*
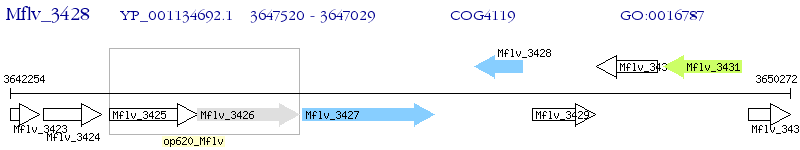
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3428 | - | - | 100% (163) | NUDIX hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3760c | - | 7e-54 | 64.19% (148) | hypothetical protein Mb3760c |
| M. tuberculosis H37Rv | Rv3733c | - | 7e-54 | 64.19% (148) | hypothetical protein Rv3733c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5270 | - | 7e-57 | 67.57% (148) | hypothetical protein MMAR_5270 |
| M. avium 104 | MAV_0356 | - | 1e-55 | 65.54% (148) | nudix hydrolase |
| M. smegmatis MC2 155 | MSMEG_3679 | - | 1e-58 | 69.59% (148) | phosphohydrolase |
| M. thermoresistible (build 8) | TH_2220 | - | 1e-60 | 62.50% (176) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4344 | - | 1e-45 | 67.50% (120) | hypothetical protein MUL_4344 |
| M. vanbaalenii PYR-1 | Mvan_3182 | - | 3e-74 | 80.37% (163) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3428|M.gilvum_PYR-GCK ---MPKLSAGLLLFRETDGVVEVLLGHPGGPFWARKDEGAWSIPKGEYAD
Mvan_3182|M.vanbaalenii_PYR-1 ---MPKLSAGLLLFRESDGVLEVLLGHPGGPFWARKDEGAWSIPKGEYTE
TH_2220|M.thermoresistible__bu ---VPKRSAGLLVYRIVDGVVEVLICHPGGPYWASKDAGVWSIPKGEYEP
Mb3760c|M.bovis_AF2122/97 ---MPKLSAGVLLYRARAGVVDVLLAHPGGPFWAGKDDGAWSIPKGEYTG
Rv3733c|M.tuberculosis_H37Rv ---MPKLSAGVLLYRARAGVVDVLLAHPGGPFWAGKDDGAWSIPKGEYTG
MAV_0356|M.avium_104 ---MPKLSAGVLLYRTGDGVVEVLIAHPGGPFWARKDDGAWSIPKGEYDE
MMAR_5270|M.marinum_M ---MPKLSAGVLVYRASAGVLELLIVHPGGPFWARKDDGAWSIPKGEYTQ
MUL_4344|M.ulcerans_Agy99 ---MPKLSAGVLVYRAAAGVLELLIVHPGGPFWARKDDGAWSIPKGEYTQ
MSMEG_3679|M.smegmatis_MC2_155 MNGMPKLSAGVLLYRVVDDVVEVLIAHPGGPFWARRDDGAWSVPKGEYTD
:** ***:*::* .*:::*: *****:** :* *.**:*****
Mflv_3428|M.gilvum_PYR-GCK GDDPWAVAQREFTEETGKPVPAGPPIGLAPVRQPGGKVVTAFAVRGDLDL
Mvan_3182|M.vanbaalenii_PYR-1 GEDPWTVAQREFTEELGKPPPTGPRIDLAAVRQPGGKVVTAFAVRGDLDL
TH_2220|M.thermoresistible__bu DEDPWAVARREFIEEVGKQPPSGPAVRFDPVRQPSGKIVTAFAVRGDVDL
Mb3760c|M.bovis_AF2122/97 GEDPWLAARREFSEEIGLCVPDGPRIDFGSLKQSGGKVVTVFGVRADLDI
Rv3733c|M.tuberculosis_H37Rv GEDPWLAARREFSEEIGLCVPDGPRIDFGSLKQSGGKVVTVFGVRADLDI
MAV_0356|M.avium_104 GEEPWPAARREFAEELGLAVPPGERIDLGTLKQSGGKLVTVFAVHGDLDV
MMAR_5270|M.marinum_M DEDPWVAARREFAEEIGLPVPDGARIDLGSVKQAGGKVVTAFAVRGDLDV
MUL_4344|M.ulcerans_Agy99 DEDPWVAARREFAEEIGLPVPDGARIDLGSVKQAGGKVVTAFAVRGDLDV
MSMEG_3679|M.smegmatis_MC2_155 DEDRWAAAQREFAEELGSPPPDGPRRELTPVRQSGGKVVTAFAVRGDFDV
.:: * .*:*** ** * * * : .::*..**:**.*.*:.*.*:
Mflv_3428|M.gilvum_PYR-GCK DGTFSNTFTLVWPRGSGTVREFPEIDRVEWFDLATARVKLLKGQRPLLDE
Mvan_3182|M.vanbaalenii_PYR-1 CDAVSNTFTLEWPRGSGIFKEFPEIDRVAWFGLAEARAKLLKGQLPLLDR
TH_2220|M.thermoresistible__bu EGAVSNTFEMEWPKGSGQIREFPEIDRVGWFPVAVAREKLVKGQRPLLDR
Mb3760c|M.bovis_AF2122/97 TDARSSTFELDWPKGSGKMRKFPEVDRVSWFPVARARTKLLKGQRGFLDR
Rv3733c|M.tuberculosis_H37Rv TDARSSTFELDWPKGSGKMRKFPEVDRVSWFPVARARTKLLKGQRGFLDR
MAV_0356|M.avium_104 TEARSNTFALEWPKGSGTLREFPEVDRVGWFPVAAARTKLLKSQHGFLDR
MMAR_5270|M.marinum_M TEAHSNTFELEWPKGSGKVREFPEVDRVGWFPVATARGKLLRGQRDFLDR
MUL_4344|M.ulcerans_Agy99 TEAHSNTFELEWPKGSGRVREFPVAG-SARHPAGPQAGSPAPKDHE----
MSMEG_3679|M.smegmatis_MC2_155 TSARSNTFEMEWPKGSGRMREFPEIDRAGWFPVAVARRKLLAGQRPLLDE
: *.** : **:*** .::** . . . . :
Mflv_3428|M.gilvum_PYR-GCK LEKV--LA------------HDGPDASRSR
Mvan_3182|M.vanbaalenii_PYR-1 LEEA--LG------------RGGPDVSRSR
TH_2220|M.thermoresistible__bu LMELPELAGCAEDQATDGPAADGPAGSRSH
Mb3760c|M.bovis_AF2122/97 LMAHPAVAG----------LSEGPESLPR-
Rv3733c|M.tuberculosis_H37Rv LMAHPAVAG----------LSEGPESLPR-
MAV_0356|M.avium_104 LMAQPAVAG----------LSEGT------
MMAR_5270|M.marinum_M LMADPSVAG----------LGEG-------
MUL_4344|M.ulcerans_Agy99 ------------------------------
MSMEG_3679|M.smegmatis_MC2_155 LMAAPELAG----------YREGD------