For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGRTSRQSPRRLLQLLMVLVAPLLVVTLGAPVAQAQPGVLVSPGMEIHQDSVLCTLGYVDPGTRIAYTAG HCRASGTVSDKFGTPIGTQGSFRDNTPDGMTVDTNHQITDWEVIHLNQDVVINNVLPSGKALVTDPSVLP VKGMPVCHFGVVTGESCGTIESVNNGWFTMANGVVSRKGDSGGPVYTPTPDGRTVLIGLFNSTWGTFPAA ISWEVASRQAQADTISAASAVVTPEPVTP*
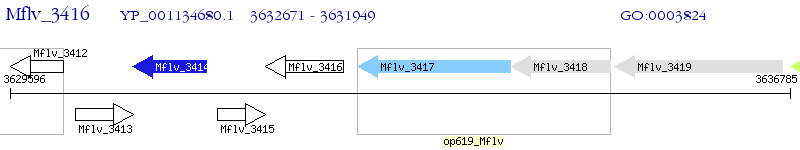
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3416 | - | - | 100% (240) | hypothetical protein Mflv_3416 |
| M. gilvum PYR-GCK | Mflv_0488 | - | 3e-05 | 27.60% (192) | hypothetical protein Mflv_0488 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1845 | - | 2e-62 | 53.99% (213) | hypothetical protein Mb1845 |
| M. tuberculosis H37Rv | Rv1815 | - | 1e-61 | 53.52% (213) | hypothetical protein Rv1815 |
| M. leprae Br4923 | MLBr_02242 | - | 6e-08 | 27.06% (170) | hypothetical protein MLBr_02242 |
| M. abscessus ATCC 19977 | MAB_2805c | - | 3e-44 | 39.82% (221) | hypothetical protein MAB_2805c |
| M. marinum M | MMAR_2692 | - | 4e-57 | 53.61% (194) | hypothetical protein MMAR_2692 |
| M. avium 104 | MAV_2901 | - | 5e-65 | 54.85% (206) | hypothetical protein MAV_2901 |
| M. smegmatis MC2 155 | MSMEG_3661 | - | 2e-75 | 60.09% (218) | hypothetical protein MSMEG_3661 |
| M. thermoresistible (build 8) | TH_2227 | - | 9e-78 | 62.26% (212) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1916 | - | 1e-26 | 31.19% (218) | hypothetical protein MUL_1916 |
| M. vanbaalenii PYR-1 | Mvan_3169 | - | 1e-112 | 81.43% (237) | hypothetical protein Mvan_3169 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1845|M.bovis_AF2122/97 ----------MVR-LVPRAFAATVALLAAGFS-PATASADPVLVFPGMEI
Rv1815|M.tuberculosis_H37Rv ----------MVR-LVPRAFAATVALLAAGFS-PATASADPVLVFPGMEI
MAV_2901|M.avium_104 ----------MHR-LATRAFAASITVTALAATLPASAAKADVMVYPGMEI
MMAR_2692|M.marinum_M ----------MVRRLALRAFAVSATTAALAFAWAPAARAEPPLVFPGMEI
Mflv_3416|M.gilvum_PYR-GCK --MSGRTSRQSPRRLLQLLMVLVAPLLVVTLGAPVAQAQPGVLVSPGMEI
Mvan_3169|M.vanbaalenii_PYR-1 --MSGRSSS----RVLHLLFAVLAPVLALALQTVPAQAQPGVLVFPGMEI
MSMEG_3661|M.smegmatis_MC2_155 --MR--------QLLSHTLRALPVFFMMAALSAWPSHADPGVQVFPGMEI
TH_2227|M.thermoresistible__bu -------------MVLAPLAAL-------TLASAPAHSSPGVVVHPGMEI
MAB_2805c|M.abscessus_ATCC_199 ------------MSAVAAVAGVLVASPALSIADPAAETAPKIVAFPGMSI
MUL_1916|M.ulcerans_Agy99 -----------MYHRWCRAAIAVVFVLVSVETPALGRAAVGVTAYPGLEI
MLBr_02242|M.leprae_Br4923 MRSIYWFTGETCATVRWLRGLGLASAAVIAPKVFASMVLAEYAPSPGLRL
**: :
Mb1845|M.bovis_AF2122/97 RQDNHVCTLGYVDP---ALKIAFTAGHCRGGG-AVTSRDYKVIGHLRAFR
Rv1815|M.tuberculosis_H37Rv RQDNHVCTLGYVDP---ALKIAFTAGHCRGGG-AVTSRDYKVIGHLRAIR
MAV_2901|M.avium_104 RQDNRVCTLGYVDP---GLKIAFTAGHCRGGGGVVTDRGGTVIGRMAAFR
MMAR_2692|M.marinum_M HQDNHVCTLAWVDP---AMHMAFTAGHCRGDG-PVTDRGNAIIGQLRAFR
Mflv_3416|M.gilvum_PYR-GCK HQDSVLCTLGYVDP---GTRIAYTAGHCRASG-TVSDKFGTPIGTQGSFR
Mvan_3169|M.vanbaalenii_PYR-1 HQDTVLCTLGYVDP---QTRIGYTAGHCRASG-TVSDKFGTMIGTQGTFR
MSMEG_3661|M.smegmatis_MC2_155 RQDSNLCTIGFIDP---VARVAFTAGHCRGSG-SVGDRNGNFIGMQAAFH
TH_2227|M.thermoresistible__bu HQDTNSCTLGFIDA---VRRIAYTAGHCRGSG-TVTDRAGRFIGTQLMFR
MAB_2805c|M.abscessus_ATCC_199 VQGDSRCTLGYVDP---VSRIGLTAGHCNATG-SIVDLAGNRLGEVLLMS
MUL_1916|M.ulcerans_Agy99 HQGSVVCVIGFVEP---QMRIAFSAGRCNGDP-MVTDRYGNAVGAVTLAG
MLBr_02242|M.leprae_Br4923 CDESSICTAACVARGNDGSSYLLTIGHCDAYDGSVWPYGPDVPLGKRTVS
: *. . : : *:* . :
Mb1845|M.bovis_AF2122/97 DNTPSGSTVATHELIADYEAIVLADDVTASNILPSGRALESRPGVVLHPG
Rv1815|M.tuberculosis_H37Rv DNTPSGSTVATHELIADYEAIVLADDVTASNILPSGRALESRPGVVLHPG
MAV_2901|M.avium_104 DNTPSGTTVSTSQVINDYEAIVLDNGVTANNILPGGRPLVSDPGLALTPG
MMAR_2692|M.marinum_M DNTPSGTTVATDQTIEDYEAIMLVDGVRISNIMPGGRPLTSTPGLALQPG
Mflv_3416|M.gilvum_PYR-GCK DNTPDGMTVDTNHQITDWEVIHLNQDVVINNVLPSGKALVTDPSVLPVKG
Mvan_3169|M.vanbaalenii_PYR-1 DNTPDGMTVDTNHQIIDWEVIHLAPHVAVNNVLPGGKVLVTDPAVLPVRG
MSMEG_3661|M.smegmatis_MC2_155 DNTPNGATVDTNHVISDWETIQLAGDVGINPALPTGRILVEDPAIGAAPG
TH_2227|M.thermoresistible__bu DNTPNGSTVATDHQIADWQTIQLAGDVVMNNVLPTGRVLVEDPTVVPQPG
MAB_2805c|M.abscessus_ATCC_199 RNLPTGGAIGPGDVVSDYEGISFGPGVELSSNLPNGLALRMAQNAGPELG
MUL_1916|M.ulcerans_Agy99 QATVSEVAVDGAAAGVDYEVITLAPEVAATDLLPTGLRLQSKPGFRVQPG
MLBr_02242|M.leprae_Br4923 ENE--GDRKDAAIILLDLSQGYSSGDVYSQLLRN---VLSENE---IQLG
* . * * *
Mb1845|M.bovis_AF2122/97 QAVCHFGVSTGETCGTVESVNNGWFTMSHGVLSEKGDSGGPVYLAPDGGP
Rv1815|M.tuberculosis_H37Rv QAVCHFGVSTGETCGTVESVNNGWFTMSHGVLSEKGDSGGPVYLAPDGGP
MAV_2901|M.avium_104 QPVCHFGVITGETCGTVESVNNGWFTMSHGVQSHNGDSGGPVYLAPSGGP
MMAR_2692|M.marinum_M QAVCHFGITTGETCGTVESVNNGWFTMTGGVESQRGDSGGPVYLAG-AGP
Mflv_3416|M.gilvum_PYR-GCK MPVCHFGVVTGESCGTIESVNNGWFTMANGVVSRKGDSGGPVYTPTPDGR
Mvan_3169|M.vanbaalenii_PYR-1 MPVCHFGVVTGESCGTIESVNNGWFTMANGVVSRKGDSGGPVYTPTPDGR
MSMEG_3661|M.smegmatis_MC2_155 MPVCHFGVVTGESCGTVEAVNNGWFTMANGVVSARGDSGGSVYTVTPDGR
TH_2227|M.thermoresistible__bu QPVCHFGVVTGESCGRIAAVNNGWFTMGDGVVSQKGDSGGPVYTVNEAGQ
MAB_2805c|M.abscessus_ATCC_199 LPVCLMGSVSGETCGKVEAINNGWFTMVD-VAGQHGDSGGPVYTVTQPGQ
MUL_1916|M.ulcerans_Agy99 ARVCEATAAR-QRCGRVDTVTESRFVIAG-MDADQHEIGAPVYVLDDDNH
MLBr_02242|M.leprae_Br4923 MLFCKVGAITGETCGTIKSMDG--DVVEASVYSLSAGSGSPGFVKNVDGI
.* : ** : :: .: : . *.. : .
Mb1845|M.bovis_AF2122/97 AQIVGIFNSVWGGFPAAVSWRSTSEQVHADLGVTPLA-------------
Rv1815|M.tuberculosis_H37Rv AQIVGIFNSVWGGFPAAVSWRSTSEQVHADLGVTPLA-------------
MAV_2901|M.avium_104 GQIVGIFNSVWGEYPAAVSWQATSEEVREDLGVRDNAR------------
MMAR_2692|M.marinum_M GQLVGIFNSIWGDFPAAVSWQRTSDEMRQDLGVPFAG-------------
Mflv_3416|M.gilvum_PYR-GCK TVLIGLFNSTWGTFPAAISWEVASRQAQADTISAASAV------------
Mvan_3169|M.vanbaalenii_PYR-1 TVLVGLFNSTWGTLPAAISWQVASQMAEADTISAASAV------------
MSMEG_3661|M.smegmatis_MC2_155 AVILGLFNSTWGQYPAAVSWPSVSGAARHTIVQASADS------------
TH_2227|M.thermoresistible__bu AVIIGMFNSTWGQFPAAVSWQSTTAQINDEIIKATAGT------------
MAB_2805c|M.abscessus_ATCC_199 ALLVGIYSLRWGGKPAAMSWSAISAQMQSQLVAQMPAAGPGNGGPELPAP
MUL_1916|M.ulcerans_Agy99 ALIAGLLDGVRGSISEAQSWPAVMRQVYLDSRSVDSTQ------------
MLBr_02242|M.leprae_Br4923 VSAVGILMSSPEGDDHTTYFVPMPPLLEKRGLQSFPEVLKR---------
*: : :
Mb1845|M.bovis_AF2122/97 ------------
Rv1815|M.tuberculosis_H37Rv ------------
MAV_2901|M.avium_104 ------------
MMAR_2692|M.marinum_M ------------
Mflv_3416|M.gilvum_PYR-GCK VTPEPVTP----
Mvan_3169|M.vanbaalenii_PYR-1 VAPQP-------
MSMEG_3661|M.smegmatis_MC2_155 ADLFVQ------
TH_2227|M.thermoresistible__bu PAAAA-------
MAB_2805c|M.abscessus_ATCC_199 PTPAAVSAAS--
MUL_1916|M.ulcerans_Agy99 PPPQGRTIGS--
MLBr_02242|M.leprae_Br4923 LSPRLVGWGSGS