For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKGIILAGGSGSRLHPITYGVSKQLIPVYDKPMVYYPLSTLMLAGIRDILVITTPHDAHSFERLLGDGSR FGVSITFAQQPSPDGLAQAFTIGEDFIGSDKVALVLGDNLLYGPGLGTQLRCFADVDGGTIFAYWVSEPS AYGVVEFDAGGLVVSLEEKPKRPKSNYAVPGLYFYDNDVVAIARDLTPSERGEYEITDVNRAYLEQGRLR VQVLPRGTAWLDTGTFDQMTDAADFVRTIERRTGLKIGVPEEIAWRQGFLSDDELCDRATRLVKSGYGRY LLDLLDRGL
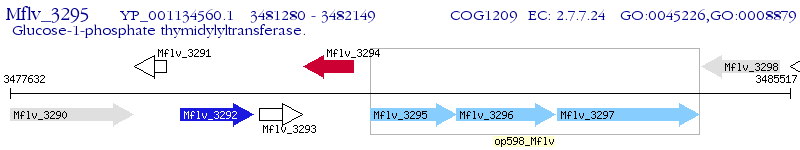
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3295 | - | - | 100% (289) | glucose-1-phosphate thymidylyltransferase |
| M. gilvum PYR-GCK | Mflv_0285 | - | e-121 | 71.43% (287) | glucose-1-phosphate thymidylyltransferase |
| M. gilvum PYR-GCK | Mflv_4734 | - | 3e-17 | 29.10% (244) | nucleotidyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0341 | rmlA | 1e-121 | 70.73% (287) | glucose-1-phosphate thymidylyltransferase |
| M. tuberculosis H37Rv | Rv0334 | rmlA | 1e-121 | 70.73% (287) | alpha-D-glucose-1-phosphate thymidylyltransferase RmlA |
| M. leprae Br4923 | MLBr_02503 | rfbA | 1e-118 | 70.14% (288) | glucose-1-phosphate thymidyltransferase |
| M. abscessus ATCC 19977 | MAB_4113 | - | 1e-125 | 72.82% (287) | glucose-1-phosphate thymidylyltransferase |
| M. marinum M | MMAR_0606 | rmlA | 1e-123 | 72.13% (287) | alpha-D-glucose-1-phosphate thymidylyl- transferase, RmlA |
| M. avium 104 | MAV_4820 | rfbA | 1e-125 | 72.13% (287) | glucose-1-phosphate thymidylyltransferase |
| M. smegmatis MC2 155 | MSMEG_0384 | rfbA | 1e-121 | 72.82% (287) | glucose-1-phosphate thymidylyltransferase |
| M. thermoresistible (build 8) | TH_1924 | rmlA | 1e-123 | 73.17% (287) | ALPHA-D-GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE RMLA |
| M. ulcerans Agy99 | MUL_0568 | rmlA | 1e-123 | 72.13% (287) | alpha-D-glucose-1-phosphate thymidylyl- transferase, RmlA |
| M. vanbaalenii PYR-1 | Mvan_0603 | - | 1e-120 | 72.47% (287) | glucose-1-phosphate thymidylyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0341|M.bovis_AF2122/97 MRGIILAGGSGTRLYPITMGISKQLLPVYDKPMIYYPLTTLMMAGIRDIQ
Rv0334|M.tuberculosis_H37Rv MRGIILAGGSGTRLYPITMGISKQLLPVYDKPMIYYPLTTLMMAGIRDIQ
MMAR_0606|M.marinum_M MRGIILAGGSGTRLYPITMGISKQLLPVYDKPMIYYPLTTLMMAGIRDIQ
MUL_0568|M.ulcerans_Agy99 MRGIILAGGSGTRLYPITMGISKQLLPVYDKPMIYYPLTTLMMAGIRDIQ
MLBr_02503|M.leprae_Br4923 MRGIILAGGSGTRLYPITLGISKQLLPVYDKPMIYYPLSTLMMAGIRDIL
MAV_4820|M.avium_104 MRGIILAGGSGTRLHPITTGISKQLLPVYDKPMVYYPLSTLMMAGIRDIL
Mvan_0603|M.vanbaalenii_PYR-1 MRGIILAGGSGTRLYPITLGISKQLVPVYDKPLVYYPLSTLMMAGIRDIL
MSMEG_0384|M.smegmatis_MC2_155 MRGIILAGGSGTRLHPLTIGVSKQLLPVYDKPLVYYPLSTLIMAGIRDIL
TH_1924|M.thermoresistible__bu MRGIILAGGSGTRLHPITMGVSKQLLPVYDKPLIYYPLSTLIMAGIRDIL
MAB_4113|M.abscessus_ATCC_1997 MRGIILAGGSGTRLHPITTGVSKQLLPVYDKPLVYYPLSTLIMAGVRDIL
Mflv_3295|M.gilvum_PYR-GCK MKGIILAGGSGSRLHPITYGVSKQLIPVYDKPMVYYPLSTLMLAGIRDIL
*:*********:**:*:* *:****:******::****:**::**:***
Mb0341|M.bovis_AF2122/97 LITTPHDAPGFHRLLGDGAHLGVNISYATQDQPDGLAQAFVIGANHIGAD
Rv0334|M.tuberculosis_H37Rv LITTPHDAPGFHRLLGDGAHLGVNISYATQDQPDGLAQAFVIGANHIGAD
MMAR_0606|M.marinum_M LITTPHDAAGFHRLLGDGTHFGVNITYATQDQPDGLAQAFVIGADHIGTD
MUL_0568|M.ulcerans_Agy99 LITTPHDAAGFHRLLGDGTHFGVNITYATQDQPDGLAQAFVIGADHIGTD
MLBr_02503|M.leprae_Br4923 VITTAHDAPGFKRLLGDGTQFGVNISYATQDHPDGLAQAFVIGANHIGAD
MAV_4820|M.avium_104 VITTPHDAAGFVRLLGDGSQFGINITYVTQERPDGLAQAFVLGADHIGND
Mvan_0603|M.vanbaalenii_PYR-1 VITTAHDAPDFRRLLGDGSDLGVNLSYAVQDEPDGLAQAFVIGASHIGTD
MSMEG_0384|M.smegmatis_MC2_155 VITTPADAPAFRRLLGDGSDFGVNLSYAAQNEPEGLAQAFLIGADHIGND
TH_1924|M.thermoresistible__bu VITTPTDAPAFQRLLGDGSAFGVNLRYAEQQEPEGLAQAFVIGADHIGDD
MAB_4113|M.abscessus_ATCC_1997 VITTPADAPAFERLLGDGSAFGINLSYAVQPQPEGLAQAFVIGAQHIGTD
Mflv_3295|M.gilvum_PYR-GCK VITTPHDAHSFERLLGDGSRFGVSITFAQQPSPDGLAQAFTIGEDFIGSD
:***. ** * ******: :*:.: :. * *:****** :* ..** *
Mb0341|M.bovis_AF2122/97 SVALVLGDNIFYGPGLGTSLKRFQSISGGAIFAYWVANPSAYGVVEFGAE
Rv0334|M.tuberculosis_H37Rv SVALVLGDNIFYGPGLGTSLKRFQSISGGAIFAYWVANPSAYGVVEFGAE
MMAR_0606|M.marinum_M SVALVLGDNIFYGPGLGTSLRRFQDISGGTIFAYWVANPTAYGVVEFSAD
MUL_0568|M.ulcerans_Agy99 SVALVLGDNIFYGPGLGTSLRRFQDISGGTIFAYWVANPTAYGVVEFSAD
MLBr_02503|M.leprae_Br4923 TVALVLGDNIFYGPGLGTSLRRFQYVSGGAIFAYCVANPSSYGIVELGID
MAV_4820|M.avium_104 SVALVLGDNIFYGPGLGTSLRRFQTITGGAVFAYWVANPSAYGVVEFSDD
Mvan_0603|M.vanbaalenii_PYR-1 SVALVLGDNIFYGPGLGTSLQRFQNVSGGAIFAYWVANPSAYGVVEFAED
MSMEG_0384|M.smegmatis_MC2_155 TVALALGDNIFYGPGLGTSLRRFEHVSGGAIFAYWVANPSAYGVVEFDAD
TH_1924|M.thermoresistible__bu TVALALGDNIFYGPGLGTSLRRFQNVTGGAIFAYWVANPSAYGVVEFGPD
MAB_4113|M.abscessus_ATCC_1997 TAMLALGDNVFYGPGLGTSLRRFENIDGGAIFAYWVANPSAYGVVEFDAA
Mflv_3295|M.gilvum_PYR-GCK KVALVLGDNLLYGPGLGTQLRCFADVDGGTIFAYWVSEPSAYGVVEFDAG
.. *.****::*******.*: * : **::*** *::*::**:**:
Mb0341|M.bovis_AF2122/97 GMALSLEEKPVTPKSNYAVPGLYFYDNDVIEIARGLKKSARGEYEITEVN
Rv0334|M.tuberculosis_H37Rv GMALSLEEKPVTPKSNYAVPGLYFYDNDVIEIARGLKKSARGEYEITEVN
MMAR_0606|M.marinum_M GTALSLEEKPATPKSHYAVPGLYFYDNDVIEIARGLKKSARGEYEITEVN
MUL_0568|M.ulcerans_Agy99 GTALSLEEKPATPKSHYAVPGLYFYDNDVIEIARSLKKSARGEYEITEVN
MLBr_02503|M.leprae_Br4923 GIALSLEEKPATPKSQYAVPGLYFYDNDVVEIARGLTKSARGEYEITEVN
MAV_4820|M.avium_104 GMALSLEEKPKTPKSNYAVPGLYFYDNDVIEIAKGLKKSERGEYEITEVN
Mvan_0603|M.vanbaalenii_PYR-1 GTALSLEEKPATPKSHYAVPGLYFYDNDVIEIARGLRRSARGEYEITEVN
MSMEG_0384|M.smegmatis_MC2_155 GKAVSLEEKPKTPKSHYAVPGLYFYDNTVIDIARSLKKSARGEYEITEVN
TH_1924|M.thermoresistible__bu GKAVSLEEKPKVPKSNYAVPGLYFYDNDVIEIARSLRKSARGEYEITEVN
MAB_4113|M.abscessus_ATCC_1997 GVPLSLEEKPATPKSHYAVPGLYFYDNDVIEIARSLQKSARGEYEITEIN
Mflv_3295|M.gilvum_PYR-GCK GLVVSLEEKPKRPKSNYAVPGLYFYDNDVVAIARDLTPSERGEYEITDVN
* :****** ***:*********** *: **:.* * *******::*
Mb0341|M.bovis_AF2122/97 QVYLNQGRLAVEVLARGTAWLDTGTFDSLLDAADFVRTLERRQGLKVSIP
Rv0334|M.tuberculosis_H37Rv QVYLNQGRLAVEVLARGTAWLDTGTFDSLLDAADFVRTLERRQGLKVSIP
MMAR_0606|M.marinum_M QAYLNHGRLNVEMLARGTAWLDTGTFDSLLDAADFVRTLERRQGLKVSVP
MUL_0568|M.ulcerans_Agy99 QAYLNHGRLNVEMLARGTAWLDTGTFDSLLDAADFVRTLERRQGLKVSVP
MLBr_02503|M.leprae_Br4923 QIYLNQGRLTVEVLARGTAWLDTGTFDSLLDASDFVRTLERRQGLKVSVP
MAV_4820|M.avium_104 QIYLSRGRLSVEVMARGTAWLDTGTFDSLLDASDFVRTLERRQGLKVSVP
Mvan_0603|M.vanbaalenii_PYR-1 QTYLNQGRLSVEVLARGTAWLDTGTFDSLLDAADFVRTIERRQGLKISVP
MSMEG_0384|M.smegmatis_MC2_155 QIYLNRGQLSVEVLARGTAWLDTGTFDSLLDASDFVRTIELRQGLKVGAP
TH_1924|M.thermoresistible__bu QIYLERGQLSVEVLARGTAWLDTGTFDSLLDASDFVRTIERRQGLKIGAP
MAB_4113|M.abscessus_ATCC_1997 QIYLDQGRLSVEVLPRGTAWLDTGTFDSLLDASDYVRTIERRQGLKIGVP
Mflv_3295|M.gilvum_PYR-GCK RAYLEQGRLRVQVLPRGTAWLDTGTFDQMTDAADFVRTIERRTGLKIGVP
: **.:*:* *:::.************.: **:*:***:* * ***:. *
Mb0341|M.bovis_AF2122/97 EEVAWRMGWIDDEQLVQRARALVKSGYGNYLLELLERN-----
Rv0334|M.tuberculosis_H37Rv EEVAWRMGWIDDEQLVQRARALVKSGYGNYLLELLERN-----
MMAR_0606|M.marinum_M EEVAWRLGWIDDEQLMRRARSLVKSGYGSYLLELLERN-----
MUL_0568|M.ulcerans_Agy99 EEVAWRLGWIDDEQLMRRARSLVKSGYGSYLLELLERN-----
MLBr_02503|M.leprae_Br4923 EEVSWRMGWIDDEQLALRAHSLAKSGYGCYLSELLERG-----
MAV_4820|M.avium_104 EEVAWQQGWITDEQLSERAQSLLKSGYGGYLLDLLERSRFH--
Mvan_0603|M.vanbaalenii_PYR-1 EEVAWRVGFIDDDALEARAKTLLKSGYGAYLLELLQR------
MSMEG_0384|M.smegmatis_MC2_155 EEIAWRAGFIDDDQLATRAKELLKSGYGHYLLQLLDRE-----
TH_1924|M.thermoresistible__bu EEIAWRAGFIDDEQLAARARQLLKSGYGTYLLELLGRERREPF
MAB_4113|M.abscessus_ATCC_1997 EEIAWRAGFINDDQLAARAQKLLKSGYGSYLLQLLQRK-----
Mflv_3295|M.gilvum_PYR-GCK EEIAWRQGFLSDDELCDRATRLVKSGYGRYLLDLLDRGL----
**::*: *:: *: * ** * ***** ** :** *