For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASMNTTGASSREIALDLYRSAAVMIVVIGHWLLSVMTYDDGEFGRDNPLVLLPWTQWLTWGFQVVPVFFA VAGFASAVSWGRRDTSASRQEWVRRRVARTLGPTAVYAIFVLTVMGALTLAGIDGSVLALGGWAVAMHLW FLAVYLMVVALTPIAVAAHRRWGLAVPAVLAVCLVLVDVIGIATGYQEIRTANYFFCWAAIYQLGIAWHD GLLGRRLLIAVAVTGAAALPLLVTWGPYPVAMIGVPGVRVENSAPPSVALLTLAMVQIGVLFALVPVLNR ALSRGRWPRLIGIANDNVMALYLWHMMPVIVVTVIAYPAGLLPQPPLGSGAWWLARLEWELVLAVVTAAL LCLLFWQRRFFAASAPTFGVPIPPGLAEAMLYVGTAACALALSLLSANGFAPDGHFPVIPVALFVTGTLL VGARPRVTARTPPVRRDQTHT*
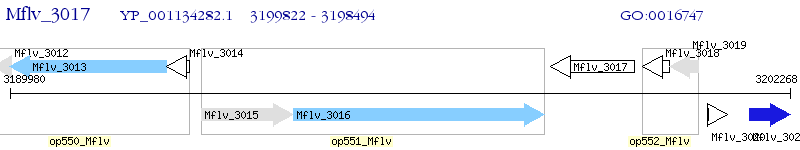
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3017 | - | - | 100% (442) | hypothetical protein Mflv_3017 |
| M. gilvum PYR-GCK | Mflv_3453 | - | 5e-36 | 32.29% (353) | hypothetical protein Mflv_3453 |
| M. gilvum PYR-GCK | Mflv_4779 | - | 2e-05 | 26.75% (314) | acyltransferase 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0233 | - | 5e-05 | 25.84% (298) | integral membrane acyltransferase |
| M. tuberculosis H37Rv | Rv0290 | - | 5e-05 | 24.48% (339) | transmembrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1297c | - | 3e-32 | 28.89% (398) | hypothetical protein MAB_1297c |
| M. marinum M | MMAR_2982 | - | 1e-131 | 53.54% (424) | hypothetical protein MMAR_2982 |
| M. avium 104 | MAV_2495 | - | 1e-127 | 53.83% (418) | hypothetical protein MAV_2495 |
| M. smegmatis MC2 155 | MSMEG_3709 | - | 6e-33 | 32.09% (349) | hypothetical protein MSMEG_3709 |
| M. thermoresistible (build 8) | TH_0475 | - | 4e-13 | 31.13% (151) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3178 | - | 4e-35 | 29.01% (424) | hypothetical protein MUL_3178 |
| M. vanbaalenii PYR-1 | Mvan_3490 | - | 0.0 | 77.01% (435) | hypothetical protein Mvan_3490 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3017|M.gilvum_PYR-GCK -------------------------MASMNTTGASS--REIALDLYRSAA
Mvan_3490|M.vanbaalenii_PYR-1 -------------------------MALMDTTRASAPERELALDLYRSSA
MMAR_2982|M.marinum_M ---MGDVIGKRRGDRARQVDHQATPTETRPNTGAKKPVRDLAVDYYRVSG
MAV_2495|M.avium_104 -----------------------------------------------MSG
MAB_1297c|M.abscessus_ATCC_199 -----------------MTMTLGLP-TADEVGAATPATRDRALDVIRIVS
TH_0475|M.thermoresistible__bu ---------------------MNFP-SPAEVAVATPATRDRAIDVIRIVS
MSMEG_3709|M.smegmatis_MC2_155 -----------------MPTPEAVPTRASDVDRATRPDRDRAVDVARLAA
MUL_3178|M.ulcerans_Agy99 -----------------MRKTSLTPDPAA-LARSTPPGRVRAVDVARLWA
Mb0233|M.bovis_AF2122/97 ----------------MGPADESGAPIRPQTPHRHTVLVTNGQVVGGTRG
Rv0290|M.tuberculosis_H37Rv MSGTVMQIVRVAILADSRLTEMALPAELPLREILPAVQRLVVPSAQNGDG
.
Mflv_3017|M.gilvum_PYR-GCK VMIVVIGHWLLSVMTYDDGEFGRDNPLVLLPWTQWLTWGFQVVPVFFAVA
Mvan_3490|M.vanbaalenii_PYR-1 VILVVIGHWLLSVMTYRDGEFGRDNPLVLMPWTQWITWGFQVVPVFFAVA
MMAR_2982|M.marinum_M VVLIVAGHWLAGSVTYHDGRFGRQNPLVELPWTQWLTWPFQAVPVFFLVA
MAV_2495|M.avium_104 VVLIVLGHWLAGSVTYHDGQFGRQNPLVDMPWTQWLTWPFQAVPTFFLVA
MAB_1297c|M.abscessus_ATCC_199 LAGVVLGHTIMAVSTIDNGVLLWGNLLNGRPVFQALTWVFQIMPLFFFAG
TH_0475|M.thermoresistible__bu LLGVVAGHTLMATSTLRDDVFIWQNLLTASPILQALTWVFQIMPLFFFAG
MSMEG_3709|M.smegmatis_MC2_155 LVAVMFGHCALLLATIDSTGVRVGNILGALPALAPITWILQVMPLFFLAG
MUL_3178|M.ulcerans_Agy99 LVVVMFGHRALLLATIDSHGVRIGNLLGELPALTPLTWLVQVMPLFFMAG
Mb0233|M.bovis_AF2122/97 FLPAVEGMRACAAVGVVVTHVAFQTGHSSGVGGRLFGRFDLAVAVFFAVS
Rv0290|M.tuberculosis_H37Rv GQADSGAAVQLSLAPVGGQPFSLDASLDTVGVVDGDLLVLQPVPAGPAAP
. . :. .
Mflv_3017|M.gilvum_PYR-GCK GFASAVSWGRRD--TSASRQEWVRRRVARTLGP-TAVYAIFVLTVMGALT
Mvan_3490|M.vanbaalenii_PYR-1 GYASAVSWARRD--AATTRQEWIRRRVARTLGP-TGVYAGFMLVVMVVLM
MMAR_2982|M.marinum_M GYAGAVSWRHHYDGDRASREAWVRHRLARVLGP-TAVYVGLISAIVVALH
MAV_2495|M.avium_104 GYAGAVSWTHRHDEEGVPRRSWLQRRLARVLGP-TAVYAVLVSAVVVALA
MAB_1297c|M.abscessus_ATCC_199 VAASVGSWKP-----GTSWGSWLMHRCTRLYRP-VFYYLGFWAVALLILR
TH_0475|M.thermoresistible__bu VAASVSGWAP-----GSSWGDWLLRRCTRLYRP-VFYYLGFWTVALVVLR
MSMEG_3709|M.smegmatis_MC2_155 GAAGTYSRH-----AGTPWGTWLFTRAQRLCRP-VFAYLAAWVVALVAVH
MUL_3178|M.ulcerans_Agy99 GAAGAYGWHPTE--PAVPWGAWLLTRAQRLCRP-VFWYLAAWTGALVVVR
Mb0233|M.bovis_AF2122/97 GFLLWRGHAA------AARDLRSHPRTGPYLRSRVARIMPAYVVAVVVIL
Rv0290|M.tuberculosis_H37Rv GIVEDIADAAMIFSTSRLKPWGIAHIQRGALAAVIAVALLATGLTVTYRV
. . :
Mflv_3017|M.gilvum_PYR-GCK LAGIDGSVLALGGWAVAMHLWFLAVYLMVVALTPIAVAAHRRWGLAVP-A
Mvan_3490|M.vanbaalenii_PYR-1 LAGIDGSVLELGGWAVAMHLWFLAVYLMVVTLTPVAVAAHQRFGLGAP-A
MMAR_2982|M.marinum_M GFGVPGSVLDYAGWAVAMHLWFLAVYVVLAALTPIAIAAHDRWGLVVP-L
MAV_2495|M.avium_104 ACGVAGSVLEYAGWAVAMHLWFLAVYLVVVSLTPIALVAQRRWGLWVP-A
MAB_1297c|M.abscessus_ATCC_199 QI-LPLHVYEPVAGVSTQLLWFLGAYVLVLAAIPLLARITTTRAMFTALA
TH_0475|M.thermoresistible__bu CL-VPVDVYRPIAGISIQLLWFLGAYVLVLAAGPXXX----GRGAVRGSA
MSMEG_3709|M.smegmatis_MC2_155 LT-LGADSTAALGRECVALLWFLGVYLVVLAFVPALTRMSSGRAVGALVT
MUL_3178|M.ulcerans_Agy99 AT-LGAQSAAGLGRECVALLWFLGVYLVVLAFVPALTRVRTGYGIATVSV
Mb0233|M.bovis_AF2122/97 SLLPDADHASLTVWLANLTLTQIYVPLTLTGGLTQMWSLSVEVAFYAALP
Rv0290|M.tuberculosis_H37Rv ATGVLAGLLAVAGIAVASALAGLLITIRSPRSGIALSIAALVPIGAALAL
* : :
Mflv_3017|M.gilvum_PYR-GCK VLAVCLVLVDVIGIATGYQEIRTANYFFCWAAIYQLGIAWHDGLLGRRLL
Mvan_3490|M.vanbaalenii_PYR-1 ALAACLVVVDVIGIGAGHPEVRILNYFFCWAAIYQLGIAWHGALVRRRLL
MMAR_2982|M.marinum_M ALAAAVVAVDAVSLGGHLPQLGWLNYLFCWGMLYQLGIAWQTGLLTGSRP
MAV_2495|M.avium_104 ALAVTVAVLDAARIGAHLPHLSWVNYVLGWGTLYQLGIAWHEGSLAGRRP
MAB_1297c|M.abscessus_ATCC_199 AVYLGIAAIDVLRLGLDAPKGIGYVNFVVWLLPAVLGVGYRRQLITQRAA
TH_0475|M.thermoresistible__bu A-------------------------------------------------
MSMEG_3709|M.smegmatis_MC2_155 ALVALAAVFDGLRFAVGTPAAGTANLVVVWLIPVVIGVAYARRLIRPRIA
MUL_3178|M.ulcerans_Agy99 TLLVLAAAVDQIRLAVGTAESGAANFLIVWLIPVALGVGYARRLIGPRAA
Mb0233|M.bovis_AF2122/97 VLALLGRRIPVGARVPAIAALAALSWAWGWLPLDAG---------SGINP
Rv0290|M.tuberculosis_H37Rv AVPGKFGPAQVLLGAAGVAAWSLIALMIPSAERERVVAFFTAAAVVGASV
.
Mflv_3017|M.gilvum_PYR-GCK IAVAVTGAAALPLLVTWGPYPVAMIGVPGVRVENSAPPSVALLTLAMVQI
Mvan_3490|M.vanbaalenii_PYR-1 VAMAVVAAAALPLLVTWGPYPIAMIGVPGDRVENSAPPSVALLALAAVQI
MMAR_2982|M.marinum_M AVLAAVSAIALTLLIWIGHYPISMIGVPGQAVQNTTPPTAAMLAFGCLQA
MAV_2495|M.avium_104 WLLAGVSAVLLAALIWLRVYPVSMIGVPGQTIDNTTPPTVALLAFGGAQT
MAB_1297c|M.abscessus_ATCC_199 LVLAATVFAINVALVTFGPYTISLVGVAGQKVPNMIPPSLVLAGHAIILS
TH_0475|M.thermoresistible__bu --------------------------------------------------
MSMEG_3709|M.smegmatis_MC2_155 LAVAVCTLAGQIMLALVGPYDVSLVVTGAERVSNVSPPTMLLALHCTWVS
MUL_3178|M.ulcerans_Agy99 LVAAVDAFAAQLRLVGTGVYDVSLVVTGADRMSNVAPPTLLLALHCTWMS
Mb0233|M.bovis_AF2122/97 LTWPPAFFSWFAAGMLLAEWAYSPVGLPHRWAR----RRVAMAVTALLGY
Rv0290|M.tuberculosis_H37Rv ALAAGAQLLWQLPLLSIGCGLIVAALLVTIQAAQLSALWARFPLPVIPAP
Mflv_3017|M.gilvum_PYR-GCK GVLFALVPVLNRALSRGRWPRLIGIANDNVMALYLWHMMPVIVVTVIAYP
Mvan_3490|M.vanbaalenii_PYR-1 GVLLALVPMLNRALARGAWPRILTVANDNVMALYLWQMLPVIVVTLIGYP
MMAR_2982|M.marinum_M GIAVMAAPALNRALRSGRVHRVLSFANNNVMALYLWHMVPVVIVAVVAYP
MAV_2495|M.avium_104 GIVMAAAPALNRALRAIRAERGLSIANNNIMALYLWHMIPVVIVAIVGYP
MAB_1297c|M.abscessus_ATCC_199 ALASAAMPAINRWAQRPRVWWAVAIGNSGAMTLYLWHMPALLGMHLAFDF
TH_0475|M.thermoresistible__bu --------------------------------------------------
MSMEG_3709|M.smegmatis_MC2_155 CLFVACARAVGRWAHRPRVWHVVAVGNGGAMTLYLWHIPVIAAAALGLHA
MUL_3178|M.ulcerans_Agy99 CAFVAAAAVIRRWAGRPRVWQLVAMGNGGAMTLYLWHIPAIAVAAFVLHA
Mb0233|M.bovis_AF2122/97 LVAASPLAGPEGLVPGTAAQFAVKTAMGSLVAFALVAPLVLDRPDTSHRL
Rv0290|M.tuberculosis_H37Rv GDPTPSAPPLRLLEDLPRRVRVSDAHQSGFIAAAVLLSVLGSVAIAVRPE
Mflv_3017|M.gilvum_PYR-GCK AGLLPQPPLGSGAWWLARLEWELVLAVVTAALLCLLFWQRRFFAASAPTF
Mvan_3490|M.vanbaalenii_PYR-1 TGLLPQPPLGSGAWWLARLEWELVLAVVTAGLLCLLYWGRRLFAAPVPTF
MMAR_2982|M.marinum_M AGLLPQPELGTTAWWLARLEWEVVLMLVTAVEMVLLWWLRRLFEAPLPTV
MAV_2495|M.avium_104 AGLLPQPPEGSVQWWQARLEWVAVLSAVTAVEMVLLWWARRLFAAPLPLL
MAB_1297c|M.abscessus_ATCC_199 LGFDRYDTTAPDFIALSVLQVATMMLLVTGLFLALRPLEN----NPLPGW
TH_0475|M.thermoresistible__bu --------------------------------------------------
MSMEG_3709|M.smegmatis_MC2_155 LGLDAFDPHAPEFWGLLALRAVVFAVLMAIAFRLLSPIEH----RPLPWW
MUL_3178|M.ulcerans_Agy99 VGLDAFDVHTPWFWCLLALRAVVFTLVMAATFWLLSPLEH----RRLPWW
Mb0233|M.bovis_AF2122/97 LGSPAMVTLG--RWSYGLFIWHLAALAMVFPVIGAFPFTG----------
Rv0290|M.tuberculosis_H37Rv ALSVVGWYLVAATAAAATLRARVWDSAACKAWLLAQPYLVAGVLLVFYTA
Mflv_3017|M.gilvum_PYR-GCK GVPIPPGLAEAMLYVGTAACALALSLLSANGFAPDGHFPVIPVALFVTGT
Mvan_3490|M.vanbaalenii_PYR-1 NVPVTPVVGEALLYAGTAACALALSLLSADGFAPGGRLPVVATVLFAGGL
MMAR_2982|M.marinum_M GVALPARWAESVMLTGAAMSAYFLWFIATEGFAPSGRFPWVAAIVFAFGL
MAV_2495|M.avium_104 GVPLPARAAEPVLLAGAALAAYTLSVVAAEGFAPDGRFPWPTALVFTAGV
MAB_1297c|M.abscessus_ATCC_199 DGGYIARPGARSAAAGTALMLAGGFTLASVVWGLKGFG-LVCWVVVLAGL
TH_0475|M.thermoresistible__bu --------------------------------------------------
MSMEG_3709|M.smegmatis_MC2_155 D-DQVQATGARSIVAGALICVAGAVLTLMAKNGLDGVVGWSALAGFVAST
MUL_3178|M.ulcerans_Agy99 D-EPVPVVGTRASASGLLVCGARVALLLVAKNGLSGAPGWVSLGCFLVAL
Mb0233|M.bovis_AF2122/97 ---RMPTVLVLTLIFGFAIAAVSYALVESPCREALRRWERRNEPISVGEL
Rv0290|M.tuberculosis_H37Rv TGRYVAAFGAVLVLAVLMLAWVVVALNPGIASPESYSLPLRRLLGLVAAG
Mflv_3017|M.gilvum_PYR-GCK LLVGARPRVTARTPPVRRDQTHT-----
Mvan_3490|M.vanbaalenii_PYR-1 LLVAARPSVVTTVPAQE-----------
MMAR_2982|M.marinum_M VLVACRP-ARAE----------------
MAV_2495|M.avium_104 LLAALRPTARPVERPA------------
MAB_1297c|M.abscessus_ATCC_199 ILARVLANRTKI----------------
TH_0475|M.thermoresistible__bu ----------------------------
MSMEG_3709|M.smegmatis_MC2_155 AAARLCAGRTGPAGTGSAARG-------
MUL_3178|M.ulcerans_Agy99 VAARAMTGPPSGAGEAQRAPAAVRQRVG
Mb0233|M.bovis_AF2122/97 QADAIAP---------------------
Rv0290|M.tuberculosis_H37Rv LDVSLIPVMAYLVGLFAWVLNR------