For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHQHLRDGHLIAVRRNRSIVVPKAFFDDHGHVVKSLPGLLVVLHDGGYTETEIVRWLFTPDPSLAIRRDG AGEEQANARPVDALHSHQAREVVRRAQAMAY
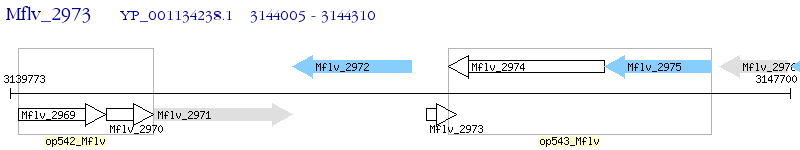
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2973 | - | - | 100% (101) | hypothetical protein Mflv_2973 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2197c | - | 2e-39 | 71.29% (101) | putative regulatory protein |
| M. tuberculosis H37Rv | Rv2175c | - | 2e-39 | 71.29% (101) | putative regulatory protein |
| M. leprae Br4923 | MLBr_00898 | - | 4e-38 | 70.30% (101) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_1990 | - | 6e-31 | 65.35% (101) | hypothetical protein MAB_1990 |
| M. marinum M | MMAR_3210 | - | 1e-40 | 72.28% (101) | regulatory protein |
| M. avium 104 | MAV_2320 | - | 6e-40 | 72.28% (101) | hypothetical protein MAV_2320 |
| M. smegmatis MC2 155 | MSMEG_4242 | - | 3e-42 | 79.21% (101) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_0710 | - | 7e-41 | 76.24% (101) | conserved hypothetical regulatory protein |
| M. ulcerans Agy99 | MUL_3518 | - | 8e-41 | 72.28% (101) | regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3538 | - | 2e-45 | 85.15% (101) | hypothetical protein Mvan_3538 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2973|M.gilvum_PYR-GCK ---------------------------------------------MHQHL
Mvan_3538|M.vanbaalenii_PYR-1 ---MQPSPRLSAMSSIPTSEDVLESDEAVYDLPTVARLLGMPVTKVHQQL
MSMEG_4242|M.smegmatis_MC2_155 ------------MSSIPAADDVLDPDEAVYDLPTVADILGIPVTKVHQQL
Mb2197c|M.bovis_AF2122/97 MPGRAPGSTLARVGSILAGDDVLDPDEPTYDLPRVAELLGVPVSKVAQQL
Rv2175c|M.tuberculosis_H37Rv MPGRAPGSTLARVGSIPAGDDVLDPDEPTYDLPRVAELLGVPVSKVAQQL
MMAR_3210|M.marinum_M ------------MGSIPAGDDVLDPDEPTYDLSRVAELLGVPVSKVHQQL
MUL_3518|M.ulcerans_Agy99 ------------MGSIPAGDDVLDPDEPTYDLSRVAELLGVPVSKVHQQL
MAV_2320|M.avium_104 ---------------------MLDPDEPIYDLPRVAELLGVPVTKVHQQL
MLBr_00898|M.leprae_Br4923 ------------MGSIPAGDDVLDPDEPTYDLTQVAELLGIPVSRVHRKL
TH_0710|M.thermoresistible__bu ------------MSSVPAADDVLDPDEPVYRLAEVAELLGIPVTKVQQQL
MAB_1990|M.abscessus_ATCC_1997 ------------MSAIPYVADTLDADEPLFTLKEVAARLRVPVSKVQQYL
: : *
Mflv_2973|M.gilvum_PYR-GCK RDGHLIAVRRNRSIVVPKAFFDDHGHVVKSLPGLLVVLHDGGYTETEIVR
Mvan_3538|M.vanbaalenii_PYR-1 REGHLVAVRRGGTIVVPKVFFDDSGHVVKSLSGLLVVLRDGGYTETEIVR
MSMEG_4242|M.smegmatis_MC2_155 RDGHLVAVRRNGVIAVPKIFFDDAGHVVKSLPGLLVVLRDGGYRTNEILR
Mb2197c|M.bovis_AF2122/97 REGHLVAVRRAGGVVIPQVFFTNSGQVVKSLPGLLTILHDGGYRDTEIMR
Rv2175c|M.tuberculosis_H37Rv REGHLVAVRRAGGVVIPQVFFTNSGQVVKSLPGLLTILHDGGYRDTEIMR
MMAR_3210|M.marinum_M REGHLVAVRREGGVVVPQVFFTSSGQVVKSLPGLLTILHDGGYHDTEIIR
MUL_3518|M.ulcerans_Agy99 REGHLVAVRREGGVVVPQVFFTSSGQVVKSLPGLLTILHDGGYHDTEIIR
MAV_2320|M.avium_104 REGHLVAVRRGEALVVPQVFFTKSGEVVKSLPGLLTILHDGGYRDTEIVR
MLBr_00898|M.leprae_Br4923 CEGYLVAVRRGDSLVVPQIFFTNSGAVVKSLPGLLTILHDGSFHETEIVR
TH_0710|M.thermoresistible__bu RDGQLVAVRRDGAPVIPKIFFDDDGQVVKNLPGLLIVLRDGGYRETEIVR
MAB_1990|M.abscessus_ATCC_1997 RDGELIAVKRDGEIKVPVIFFSPEGPVVKHLFGLLSVLRDGGYHEPEILR
:* *:**:* :* ** * *** * *** :*:**.: **:*
Mflv_2973|M.gilvum_PYR-GCK WLFTPDPSLAIRRDGAGEEQANARPVDALHSHQAREVVRRAQAMAY
Mvan_3538|M.vanbaalenii_PYR-1 WLFTPDASLTIRRDGSTEGQANARPVDALHSHQAREVVRRAQAMAY
MSMEG_4242|M.smegmatis_MC2_155 WLFTPDESLTITRDGSTERLTNARPVDALHSHQAREVIRRAQAMAY
Mb2197c|M.bovis_AF2122/97 WLFTPDPSLTITRDGSRDAVSNARPVDALHAHQAREVVRRAQAMAY
Rv2175c|M.tuberculosis_H37Rv WLFTPDPSLTITRDGSRDAVSNARPVDALHAHQAREVVRRAQAMAY
MMAR_3210|M.marinum_M WLFTPDPSLTVTRDGSRDAVSNARPVDALHAHQAREVVRRAQAMAY
MUL_3518|M.ulcerans_Agy99 WLFTPDPSLTVTRDGSRDAVSNARPVDALHAHQAREVVRRAQAMAY
MAV_2320|M.avium_104 WLFTRDPSLTVTRDGSRDAISNARPVDALHAHQAREVVRRAQAMAY
MLBr_00898|M.leprae_Br4923 WLFTPDPSLTLTRDGSRDVVSNARPVDALHTHQAREVVRRAQAMAY
TH_0710|M.thermoresistible__bu WLFTPDPSLTITRDGSREPLSNVRPVDALHSHQAREVVRRAQAMAY
MAB_1990|M.abscessus_ATCC_1997 WLFAGDESLTVSRDGTTERIEAARPVDALHGHQAREVLRRAQAMAY
***: * **:: ***: : .******* ******:********