For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRSVPVRLYEQPWPEGDFRFFQLGHVVDDVVAAAARWARTFGIGPFHVLPTVEQHTDYGGQIRPVRIQVA VAQAGPVQIELIQQHCDTPSIYSEWSRGGTSSFHQIATLTSGYDAKTAHFAALGYPIAAQSHGGGFRVAY IDTVADFGFYTEVVDAPPAFTDHVRAIAATCADWDGSDPVRILTRDGYRVP*
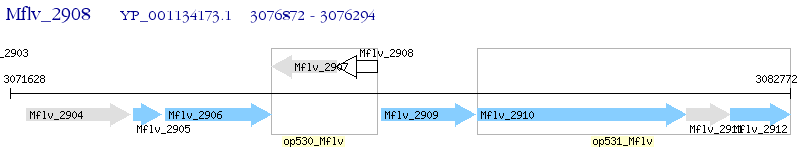
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2908 | - | - | 100% (192) | hypothetical protein Mflv_2908 |
| M. gilvum PYR-GCK | Mflv_2521 | - | 2e-18 | 29.27% (164) | hypothetical protein Mflv_2521 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2388 | - | 2e-07 | 26.71% (161) | hypothetical protein MAB_2388 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4776 | - | 2e-19 | 33.13% (163) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4137 | - | 7e-23 | 34.13% (167) | hypothetical protein Mvan_4137 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4776|M.smegmatis_MC2_155 -----MNPVLPG------------SICQIGYVVADLDDAISRWL-QMGVG
Mvan_4137|M.vanbaalenii_PYR-1 -----MT--LPG------------PIRQIGYVVTDLDRALDSWV-ALGVG
Mflv_2908|M.gilvum_PYR-GCK -----MPRSVPVRLYEQPWPEGDFRFFQLGHVVDDVVAAAARWARTFGIG
MAB_2388|M.abscessus_ATCC_1997 MTTSAMPPDVSGFG----------RVTQIAWVTDDLDATEAMLSAVFGVK
* :. . *:. *. *: : :*:
MSMEG_4776|M.smegmatis_MC2_155 PWFVIRGMTQR---VRFRGEPCEVTLSLALSNSGDLQIELIQQHGDTHSV
Mvan_4137|M.vanbaalenii_PYR-1 PWYVIREMPQR---VTYRGEPCAVTLSLALANSGDLQVELIHQHDDTPSI
Mflv_2908|M.gilvum_PYR-GCK PFHVLPTVEQH---TDYGGQIRPVRIQVAVAQAGPVQIELIQQHCDTPSI
MAB_2388|M.abscessus_ATCC_1997 KWTRIPDVHFGPETCTYRGAPADFTATIALSYLDDMQLELILPVSGT-SI
: : : : * . :*:: . :*:*** .* *:
MSMEG_4776|M.smegmatis_MC2_155 FTEFLAATGGGFHQLAYWARDFDAAVADLEDAGWPEVWSGGEAEG---VR
Mvan_4137|M.vanbaalenii_PYR-1 FTEFLDSGRPGFHQLAYWTDDFEGTMGSVAEAGWPVVWNGASGPGGAVTR
Mflv_2908|M.gilvum_PYR-GCK YSEWSRGGTSSFHQIATLTSGYDAKTAHFAALGYPIAAQS-HGGG---FR
MAB_2388|M.abscessus_ATCC_1997 YTEFLERNSAGLHHVCVEPPDLDVALDHATSQGMEVVQDGVMPGG---MR
::*: .:*::. . . : * . .. * *
MSMEG_4776|M.smegmatis_MC2_155 FAYYEPPAG-ATVIEIMELTDASAGMATFVRQAVRAWDGSDPVRELGGA-
Mvan_4137|M.vanbaalenii_PYR-1 FAYVEPPDSPAAVIEIMELSPATEGMATFLREAAAAWDGRDPVRELSV--
Mflv_2908|M.gilvum_PYR-GCK VAYIDTVADFGFYTEVVDAPPAFTDHVRAIAATCADWDGSDPVRILTRDG
MAB_2388|M.abscessus_ATCC_1997 FAYLSAPAAALPYMELVFVPPEIRAFYDYIKNEQA---------------
.** .. *:: . :
MSMEG_4776|M.smegmatis_MC2_155 ----
Mvan_4137|M.vanbaalenii_PYR-1 ----
Mflv_2908|M.gilvum_PYR-GCK YRVP
MAB_2388|M.abscessus_ATCC_1997 ----