For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADLSGPLAGSPLETSVLESLDMLERALSLQPTGHHRFRAGNERDRFGRLFGGQLLAQAVYAAARTVESH PPHSVHAYFVQTGASDTPVDIAVDVVRDGRSMATRQVTIAQGERTLLTAMASFHTNPDEPELARPHSEVP APEDVPLLQHWVPRAGAEFGAHAQNWIDVPPALEMRIAEPTGFLGGDQTPDPRSHWMRLPRPIADDPALH TAMLAYASDYLLLDMALRNHPHRVDYSSIAAVSLDHSLWVHRPVRFDDWHLYTQETVALAGHRALIRGTI RDLGGHTVASTSEEVLIRPTGARTSPMS*
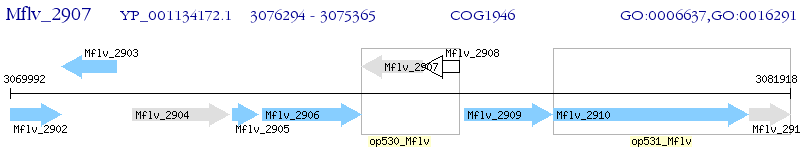
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2907 | - | - | 100% (309) | acyl-CoA thioesterase |
| M. gilvum PYR-GCK | Mflv_3831 | - | 4e-30 | 33.57% (283) | acyl-CoA thioesterase II |
| M. gilvum PYR-GCK | Mflv_1022 | - | 3e-28 | 29.27% (287) | acyl-CoA thioesterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2637c | tesB2 | 1e-29 | 33.86% (251) | acyl-CoA thioesterase II |
| M. tuberculosis H37Rv | Rv1618 | tesB1 | 1e-29 | 29.35% (293) | acyl-CoA thioesterase II TesB1 |
| M. leprae Br4923 | MLBr_01278 | tesB | 8e-27 | 27.36% (296) | acyl CoA thioesterase II |
| M. abscessus ATCC 19977 | MAB_2891c | - | 5e-31 | 35.14% (276) | acyl-CoA thioesterase II TesB2 |
| M. marinum M | MMAR_2095 | tesB2 | 2e-28 | 34.12% (255) | acyl-CoA thioesterase II TesB2 |
| M. avium 104 | MAV_2540 | - | 2e-28 | 29.30% (256) | acyl-CoA thioesterase |
| M. smegmatis MC2 155 | MSMEG_3812 | - | 2e-31 | 29.45% (275) | acyl-CoA thioesterase |
| M. thermoresistible (build 8) | TH_3198 | - | 1e-103 | 61.67% (287) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_3249 | tesB2 | 2e-28 | 34.12% (255) | acyl-CoA thioesterase II TesB2 |
| M. vanbaalenii PYR-1 | Mvan_2565 | - | 1e-28 | 31.93% (285) | acyl-CoA thioesterase II |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2907|M.gilvum_PYR-GCK MVADLSGPLAGSPLETSVLESLDMLERALSLQPTGHHRFRAGNERDRFGR
TH_3198|M.thermoresistible__bu ----MTDTVDVDHLQRSIQDTLTELLRALELEPVAADRFRAGSESDRFGR
MMAR_2095|M.marinum_M -------------------MAIEEILDLEQLEVNIYRGSVFSPESGFLQR
MUL_3249|M.ulcerans_Agy99 -------------------MAIEEILDLEQLEVNIYRGSVFSPESGFLQR
Mb2637c|M.bovis_AF2122/97 -------------------MSIEEILDLEQLEVNIYRGSVFSPESGFLQR
Mvan_2565|M.vanbaalenii_PYR-1 -------------------MAIEEILDLEQIEVNIYRGGVFSPESGFLQR
MAB_2891c|M.abscessus_ATCC_199 ------------------MADIEEILTLEQLEKDIFRGNVTPSN---LRR
MAV_2540|M.avium_104 -----------------MISTLEDVLSLLDLQQIDDAAFVGTQPDTPNHH
MSMEG_3812|M.smegmatis_MC2_155 ----------------MPLPQLADVLATLDITDCGHDMFVATQIDNEAHH
Rv1618|M.tuberculosis_H37Rv ------------MPDGKPMSDFDELLAVLDLNAVASDLFTGSHPSKNPLR
MLBr_01278|M.leprae_Br4923 ---------MKRLWEKKTMSDFEELLAVLDLNRVADDLFIGSHPSKNPLR
: : .: :
Mflv_2907|M.gilvum_PYR-GCK LFGGQLLAQAVYAAARTVE-SHPPHS-VHAYFVQTGASDTPVDIAVDVVR
TH_3198|M.thermoresistible__bu IFGGQVVGQALLAAARTVP-DHPPHS-LHAYFVQTGDGTRPLDIAVQRIR
MMAR_2095|M.marinum_M TFGGHVAGQSLVSAVRTVDPRYQVHS-LHGYFLRSGDAQEPTVFLVERTR
MUL_3249|M.ulcerans_Agy99 TFGGHVAGQSLVSAVRTVDPRYQVHS-LHGYFLRSGDAQEPTVFLVERTR
Mb2637c|M.bovis_AF2122/97 TFGGHVAGQSLVSAVRTVDPRYMVHS-LHGYFLRPGDAKERTVFLVERIR
Mvan_2565|M.vanbaalenii_PYR-1 TFGGHVAGQSLVSAVRTVDPKFQVHS-LHGYFLRAGDARSPSVYTVERIR
MAB_2891c|M.abscessus_ATCC_199 TFGGQVAGQSLVSAVRTVEPEYLVHS-LHGYFLRPGNPNEPTVFLVDRVR
MAV_2540|M.avium_104 IIGSQVAAQALMAAGRTTP-GRLAHS-MHMYFLRRGDARQPIQYDVTPLR
MSMEG_3812|M.smegmatis_MC2_155 IIGGHISAQALMAASRTAA-GRVPNS-LHVYLLRAGDARYPVEMQVMNMR
Rv1618|M.tuberculosis_H37Rv TFGGQLMAQSFVASSRTLTRHHLPPSAFSVHFINGGDTAKDIEFQVIRLR
MLBr_01278|M.leprae_Br4923 TFGGQLVAQSFVASSRTLIRDDLPPSAFSVHFINGGDTAKEIEFHVVRLR
:*.:: .*:. :: ** * . :::. * * *
Mflv_2907|M.gilvum_PYR-GCK DGRSMATRQVTIAQGERTLLTAMASFHTNPDEPELARPHSEVPAPEDVPL
TH_3198|M.thermoresistible__bu DGRSMATRQVTVTQDDRTLLTALVSFHDNPDEPRWAAPAPVLPGPEQLPV
MMAR_2095|M.marinum_M DGGSFVTRRVNAVQHGEVIFSMGASFQTAQNGISHQDAMPAAPPPDDLPG
MUL_3249|M.ulcerans_Agy99 DGGSFVTRRVNAVQHGEVIFSMGASFQTAQNGISHQDAMPAAPPPDDLPG
Mb2637c|M.bovis_AF2122/97 DGGSLCTRRVNAVQHGETIFSMAASFQTEQEGITHQDVMPAAPPPDGLPG
Mvan_2565|M.vanbaalenii_PYR-1 DGGSFCTRRVTAIQHGETIFSMSASFQTDQSGIEHQDAMPFAPPPDDIPD
MAB_2891c|M.abscessus_ATCC_199 DGRSFVTRRVTAIQDGQAIFSMSASFQTLDTGIEHQDLMPPVPDPEDLPD
MAV_2540|M.avium_104 DGGTISSRRVTASQSGVVLFEALASFTIIADDVDWQQRMPDVAGPSAVHG
MSMEG_3812|M.smegmatis_MC2_155 DGGMLSTRRVIARQGDEVLLEALVSLSDTADTVDYHQPMPDVPAPEDLPS
Rv1618|M.tuberculosis_H37Rv DERRFANRRVDAVQDGTLLSSAMVSYMAGGRGHEHALDPPQVAEPHTRPP
MLBr_01278|M.leprae_Br4923 DERRFANRRVDAIQDGMLLSSAMASYMSGGCGLEHAVEPPEAAEPHTRPP
* : .*:* * : .* . . *
Mflv_2907|M.gilvum_PYR-GCK LQHWVPRAGAEFGAHAQNWIDVPPALEMRIAEPTGFLGGDQTPDPR---S
TH_3198|M.thermoresistible__bu LQHWAATAPPALRPQVRHWIDVPPPLDIRIGEATYFLGGRPAEGPR---S
MMAR_2095|M.marinum_M LR--------SVRVFDDAGFRQFEEWDVRIVPRDLLAPLPGKASQQ---Q
MUL_3249|M.ulcerans_Agy99 LR--------SVRVFDDAGFRQFEEWDVRIVPRDLLAPLPGKASQQ---Q
Mb2637c|M.bovis_AF2122/97 LN--------SIKVFDDAGFRQFDEWDVCIVPRERLRLLPGKASQQ---Q
Mvan_2565|M.vanbaalenii_PYR-1 FKS-------SSKVFDDASFRQFDEWDVRIVPRDRLGVREGKASQQ---Q
MAB_2891c|M.abscessus_ATCC_199 AQ--------EQDSFNRDLFRQFAEWDIRIVPDELMDRNPRLAAQQ---R
MAV_2540|M.avium_104 LEDLLA----PYAEEFGDWWPQQRPFTMRYLDAPPRVALDLSDPPPPRLR
MSMEG_3812|M.smegmatis_MC2_155 VGELLS----DYADELDGFWVKPQWIERRYVDAPPRLALDQPEPPE-RTR
Rv1618|M.tuberculosis_H37Rv IGELLR----GYEETVPHFVNALQPIEWRYANDPAWIMRDKGDRLAYN-R
MLBr_01278|M.leprae_Br4923 IGELLR----GYEQIVQHFVNALQPVEWRYVNDPSWVMRNKGQRLAYN-R
Mflv_2907|M.gilvum_PYR-GCK HWMRLPRPIADDPALHTAMLAYASDYLLLDMALRNHPHRVDYSSIAAVSL
TH_3198|M.thermoresistible__bu HWMRLPRDVGDDPLLHRVLLAYASDYLLLDAALRAHPDLSGHDGFQAVSL
MMAR_2095|M.marinum_M VWFRHRDPLPDDPVLHICALAYMSDLTLLGSAQ--VTHLAEREHLQVASL
MUL_3249|M.ulcerans_Agy99 VWFRHRDPLPDDPVLHICALAYMSDLTLLGSAQ--VTHLAEREHLQVASL
Mb2637c|M.bovis_AF2122/97 VWLRHRDPLPDDPVLHICALAYMSDLTLLGSAQ--VNHLDVRDQLQVASL
Mvan_2565|M.vanbaalenii_PYR-1 VWFRHRDPMPDDHVLHICALAYMSDLTLLGSAQ--VNHIEERKHLMVASL
MAB_2891c|M.abscessus_ATCC_199 VWFRCRKHLPDDPVLHICALAYMSDLTLLSSSK--VPHRDT--VLQTASL
MAV_2540|M.avium_104 IWLRANGEVTDDPLVNSCVVAYLSALTLLECVMTTMRTTPVGPRLSAL-V
MSMEG_3812|M.smegmatis_MC2_155 MWWRPAESIPNDPVLNCCVLTYASGTSLLETTVT-MRRTTQATAFSAL-I
Rv1618|M.tuberculosis_H37Rv VWVKALGEMPDDPVLHTATLLYSSDTTVLDSVITTHGLSWGFDRIFAASA
MLBr_01278|M.leprae_Br4923 VWVKALGALPNDPTLHTAAMLYSTDTTVLDSVITTHGLSWGFDRIFAVSA
* : : :* :: : * : :* : .
Mflv_2907|M.gilvum_PYR-GCK DHSLWVHRPVRFDDWHLYTQETVALAGHRALIRGTIRDLGGHTVASTSEE
TH_3198|M.thermoresistible__bu DHSVWLHRPVRFDRWHVYTQNTTVVSGHRALVRGTIHDAAGALVASTSQE
MMAR_2095|M.marinum_M DHAMWFMRGFRADEWLLYDQSSPSAGGGRALTHGKIFTQGGELVAAVMQE
MUL_3249|M.ulcerans_Agy99 DHAMWFMRGFRADEWLLYDQSSPSAGGGRALTHGKIFTQGGELVAAVMQE
Mb2637c|M.bovis_AF2122/97 DHAMWFMRPFRADEWLLYDQSSPSASGGRALTRGEIFTRSGEMVAAVMQE
Mvan_2565|M.vanbaalenii_PYR-1 DHAMWFMRAFRADEWLLYDQSSPSACGGRSLTQGKIYNRYGEMVAAVMQE
MAB_2891c|M.abscessus_ATCC_199 DHALWFLRPFRADEWMLYDETSPSAGFGRALTQGRIFNRDGTMVAAVVQE
MAV_2540|M.avium_104 DHTIWFHRAADFTDWLLFDQFSPSIVGRRGLATGTLYNRSGELVCIATQE
MSMEG_3812|M.smegmatis_MC2_155 DHTVWFQRPADLSDWVLSDQVSLSGIHGRGLASGTMYNRSGELVCSATQE
Rv1618|M.tuberculosis_H37Rv NHSVWFHRQVNFDDWVLYSTSSPVAADSRGLGSGHFFDRSGKLIATVVQE
MLBr_01278|M.leprae_Br4923 NHSVWFHRQVNFDDWVLYSTSSPVAADSRGLGTGHLFDRSGQLIATVVQE
:*::*. * * : : *.* * : * :. . :*
Mflv_2907|M.gilvum_PYR-GCK VLIRPTGARTSPMS------
TH_3198|M.thermoresistible__bu VLIRPAER------------
MMAR_2095|M.marinum_M GLTRYPSGYQPGEK------
MUL_3249|M.ulcerans_Agy99 GLTRYPSGYQPGEN------
Mb2637c|M.bovis_AF2122/97 GLTRHRRGHRSVGQ------
Mvan_2565|M.vanbaalenii_PYR-1 GLTRYTRGYTPGHG------
MAB_2891c|M.abscessus_ATCC_199 GLTRIERNPEQASVARGNMA
MAV_2540|M.avium_104 GYFAEQRQ------------
MSMEG_3812|M.smegmatis_MC2_155 IYFGRGRI------------
Rv1618|M.tuberculosis_H37Rv GVLKYFPATPDSAAGRS---
MLBr_01278|M.leprae_Br4923 GVLKYFPASHR---------