For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KLAAVRRTAPEVLITAKTQRWTPEEVLRTLVETELAARDASNVVNRLKTAAFPVPKTLESFEVAASSIQP KVFDYLSSLEWIRTQQNLAIIGPAGTGKSHTLIGLGTAAIHAGHKVRYFTAADLVETLYRGLADNTVGKI IESLLRVDLIILDELGFAPLDDTGTQLLFRLVAGAYERRSLAIGSHWPFEQWGRFLPEQTTAVSILDRLL HHATVVITDGDSYRMKDAKHRKEQPKPT*
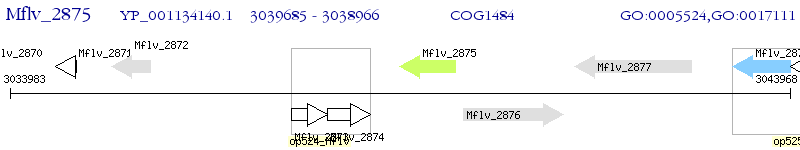
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2875 | - | - | 100% (239) | IstB ATP binding domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0579 | - | 1e-26 | 31.94% (191) | IstB ATP binding domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0693 | - | 1e-07 | 32.63% (95) | putative transposition helper protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3662 | - | 1e-30 | 31.00% (229) | transposase |
| M. tuberculosis H37Rv | Rv3638 | - | 1e-30 | 31.00% (229) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3532 | - | 1e-105 | 77.64% (237) | helper protein |
| M. smegmatis MC2 155 | MSMEG_2150 | - | 2e-37 | 34.93% (229) | putative transposition helper protein |
| M. thermoresistible (build 8) | TH_1648 | - | 9e-38 | 35.00% (240) | putative transposition helper protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3895 | - | 2e-44 | 92.63% (95) | hypothetical protein Mvan_3895 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2875|M.gilvum_PYR-GCK ------------------------------------MKLAAVRRTAPEVL
MAV_3532|M.avium_104 ----------MTTAAKPVAPSSAAPLAADLDAALRRLKLATVRRNAAEVL
Mvan_3895|M.vanbaalenii_PYR-1 -----MTKKPSITDTEPVAPPSVPPLAADLDAGLRRLKLAAVRRTAPEVL
Mb3662|M.bovis_AF2122/97 --------------MAAKTATNSRDVAAELAYLTRALKAPTLRGAIEQLA
Rv3638|M.tuberculosis_H37Rv --------------MAAKTATNSRDVAAELAYLTRALKAPTLRGAIEQLA
MSMEG_2150|M.smegmatis_MC2_155 MTPTPRTAKN-TATVTEEPPSTAASRYQKLRSHLAELKLHAAAAALPAVL
TH_1648|M.thermoresistible__bu MTPTPRTPKTATTTTVEESPSAAASRYQQLRSHLAELKLAAAAEALPAVL
:* : :
Mflv_2875|M.gilvum_PYR-GCK ITAKTQRWTPEEVLRTLVETELAARDASNVVNRLKTAAFPVPKTLESFEV
MAV_3532|M.avium_104 QVAKTQRWTPEEILRTLVEAEIAARDASNTANRLKAAAFPVTKTLDGFDV
Mvan_3895|M.vanbaalenii_PYR-1 ITAKTQRWTPEEVLRTLIETELAARDASNVVNRLKAAAFPVPKNLESFDV
Mb3662|M.bovis_AF2122/97 DRARTKTWSYEEFLAACLQREVSARESHGGEGRIRAARFPSRKSLEEFDF
Rv3638|M.tuberculosis_H37Rv DRARTKTWSYEEFLAACLQREVSARESHGGEGRIRAARFPSRKSLEEFDF
MSMEG_2150|M.smegmatis_MC2_155 DQASAENLSLTVALERLLAVEVEASTARRLAGRLRFACLPTPATLDDFDV
TH_1648|M.thermoresistible__bu DEATAEGLSLTVALERLLAVEVEASTARRLAGRLRFACLPTPATLADFDV
* :: : * : *: * : .*:: * :* .* *:.
Mflv_2875|M.gilvum_PYR-GCK AASS-IQPKVFDYLSSLEWIRTQQNLAIIGPAGTGKSHTLIGLGTAAIHA
MAV_3532|M.avium_104 TGSS-ITAATFDYLSSLEWIRAQQNLAVIGPPGTGKSHLLIGCGHAAVHA
Mvan_3895|M.vanbaalenii_PYR-1 AASS-IPPKTFDYLSSLEWIRAQQNLAIIGP---------------AVIN
Mb3662|M.bovis_AF2122/97 DHARGLKRDTIAHLGTLDFVT-----------------LAIGIAIRACQA
Rv3638|M.tuberculosis_H37Rv DHARGLKRDTIAHLGTLDFVT-----------------LAIGIAIRACQA
MSMEG_2150|M.smegmatis_MC2_155 DAAAGIDRKLIDELGTCRYLESATNILLVGPPGTGKTHLSVGLARAAAHA
TH_1648|M.thermoresistible__bu DAAAGIDRKLIDELGTCRYLESATNILLIGPPGTGKTHLSVGLARAAAHA
: : : *.: :: *
Mflv_2875|M.gilvum_PYR-GCK GHKVRYFTAADLVETLYRGLADNTVGKIIESLLRVDLIILDELGFAPLDD
MAV_3532|M.avium_104 GFKVRYFTAADLIEVLYRGLADNTVGKIIDTLLRADLVILDEIGFAPLDD
Mvan_3895|M.vanbaalenii_PYR-1 G-------------------------------------------------
Mb3662|M.bovis_AF2122/97 GHRVLFATASQWVDRLAAAHHSGTLQSELIRLARYPLLVVDEVGYIPFEP
Rv3638|M.tuberculosis_H37Rv GHRVLFATASQWVDRLAAAHHSGTLQSELIRLARYPLLVVDEVGYIPFEP
MSMEG_2150|M.smegmatis_MC2_155 GYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGPTLLVIDELGYLPLPA
TH_1648|M.thermoresistible__bu GYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGPTLLVIDELGYLPLPA
*
Mflv_2875|M.gilvum_PYR-GCK TGTQLLFRLVAGAYERRSLAIGSHWPFEQWGRFLPEQTTAVSILDRLLHH
MAV_3532|M.avium_104 TGTQLLFRLVAAGYERRSLAIASHWPFEQWGRFLPEHTTAASILDRLLHH
Mvan_3895|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3662|M.bovis_AF2122/97 EAANLFFQLVSSRYERASLIVTSNKPFGRWGEVFGDDVVAAAMIDRLVHH
Rv3638|M.tuberculosis_H37Rv EAANLFFQLVSSRYERASLIVTSNKPFGRWGEVFGDDVVAAAMIDRLVHH
MSMEG_2150|M.smegmatis_MC2_155 EAASALFQVVSQRYLKTSIVITTNRGVGSWGEILGDTTVAAAMLDRLLHR
TH_1648|M.thermoresistible__bu EAASALFQVVSQRYLKTSIVITTNRGVGAWGEILGDTTVAAAMLDRLLHR
Mflv_2875|M.gilvum_PYR-GCK ATVVITDGDSYRMKDAKHRKEQPKPT---------
MAV_3532|M.avium_104 ASIVVTSGESYRMRHADHKKGAAKN----------
Mvan_3895|M.vanbaalenii_PYR-1 -----------------------------------
Mb3662|M.bovis_AF2122/97 AEVIALKGDSYRIKD-----RDLGRVPTVTADDQ-
Rv3638|M.tuberculosis_H37Rv AEVIALKGDSYRIKD-----RDLGRVPTVTADDQ-
MSMEG_2150|M.smegmatis_MC2_155 SVVINLDGESYRLRDHQAAAETLRRATTGTRQALH
TH_1648|M.thermoresistible__bu SVVINLDGESYRLRDHQAAAETLRRTTTGTRQPLH