For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPTPRTAKNTATVTEEPPSTAASRYQKLRSHLAELKLHAAAAALPAVLDQASAENLSLTVALERLLAVEV EASTARRLAGRLRFACLPTPATLDDFDVDAAAGIDRKLIDELGTCRYLESATNILLVGPPGTGKTHLSVG LARAAAHAGYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGPTLLVIDELGYLPLPAEAASALFQVVSQ RYLKTSIVITTNRGVGSWGEILGDTTVAAAMLDRLLHRSVVINLDGESYRLRDHQAAAETLRRATTGTRQ ALH*
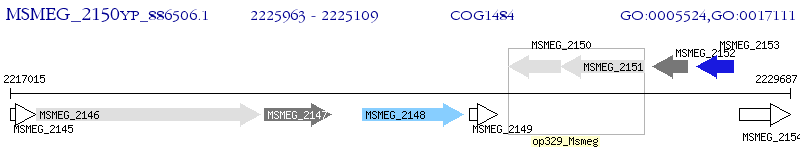
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2150 | - | - | 100% (284) | putative transposition helper protein |
| M. smegmatis MC2 155 | MSMEG_1858 | - | e-158 | 100.00% (284) | putative transposition helper protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3662 | - | 4e-35 | 36.68% (229) | transposase |
| M. gilvum PYR-GCK | Mflv_0693 | - | 2e-57 | 86.92% (130) | putative transposition helper protein |
| M. tuberculosis H37Rv | Rv3638 | - | 4e-35 | 36.68% (229) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1806 | - | 1e-71 | 87.82% (156) | LprM protein |
| M. thermoresistible (build 8) | TH_1648 | - | 1e-147 | 93.33% (285) | putative transposition helper protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5932 | - | 1e-42 | 73.04% (115) | hypothetical protein Mvan_5932 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2150|M.smegmatis_MC2_155 MTPTPRTAKN-TATVTEEPPSTAASRYQKLRSHLAELKLHAAAAALPAVL
TH_1648|M.thermoresistible__bu MTPTPRTPKTATTTTVEESPSAAASRYQQLRSHLAELKLAAAAEALPAVL
MAV_1806|M.avium_104 -------------------------------------------------M
Mflv_0693|M.gilvum_PYR-GCK MTPTP--RTKKTTTTTDETPSTSASRYQQLRSHLAALNLHACAEALPAVL
Mvan_5932|M.vanbaalenii_PYR-1 MTTTP--DAVPAPNT-----AAQASLYQRLRGHLAVLKLHDAAEALPSVL
Mb3662|M.bovis_AF2122/97 --------------MAAKTATNSRDVAAELAYLTRALKAPTLRGAIEQLA
Rv3638|M.tuberculosis_H37Rv --------------MAAKTATNSRDVAAELAYLTRALKAPTLRGAIEQLA
MSMEG_2150|M.smegmatis_MC2_155 DQASAENLSLTVALERLLAVEVEASTARRLAGRLRFACLPTPATLDDFDV
TH_1648|M.thermoresistible__bu DEATAEGLSLTVALERLLAVEVEASTARRLAGRLRFACLPTPATLADFDV
MAV_1806|M.avium_104 DQASAEGLSLTVALERLLAVEVEASTARRLAGRLRFACLPTPATLADFDV
Mflv_0693|M.gilvum_PYR-GCK DQASAEGLSLTVALERLLAVEVEASTARRLAGRLRFACIPTPATLADFDV
Mvan_5932|M.vanbaalenii_PYR-1 DRAQADGTSMTAALEELLSIEVEATEARRLAGRLRFACLPTPASLEDFDY
Mb3662|M.bovis_AF2122/97 DRARTKTWSYEEFLAACLQREVSARESHGGEGRIRAARFPSRKSLEEFDF
Rv3638|M.tuberculosis_H37Rv DRARTKTWSYEEFLAACLQREVSARESHGGEGRIRAARFPSRKSLEEFDF
*.* :. * * * **.* :: **:* * :*: :* :**
MSMEG_2150|M.smegmatis_MC2_155 DAAAGIDRKLIDELGTCRYLESATNILLVGPPGTGKTHLSVGLARAAAHA
TH_1648|M.thermoresistible__bu DAAAGIDRKLIDELGTCRYLESATNILLIGPPGTGKTHLSVGLARAAAHA
MAV_1806|M.avium_104 DAAAGIDRKLIDELGTCRYLESATNILLIGPPGTGKTHLSVGLARAAAHA
Mflv_0693|M.gilvum_PYR-GCK DAAAGIDRKLIDELGTCRYLESATNILLIGPG------------------
Mvan_5932|M.vanbaalenii_PYR-1 DAAPGLDRKLITELGTCRYLETATNVLLIGPP------------------
Mb3662|M.bovis_AF2122/97 DHARGLKRDTIAHLGTLDFVTLAIGIAIRACQAG----------------
Rv3638|M.tuberculosis_H37Rv DHARGLKRDTIAHLGTLDFVTLAIGIAIRACQAG----------------
* * *:.*. * .*** :: * .: : .
MSMEG_2150|M.smegmatis_MC2_155 GYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGPTLL-VIDELGYLPLP
TH_1648|M.thermoresistible__bu GYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGPTLL-VIDELGYLPLP
MAV_1806|M.avium_104 GYRTYFTTAADLAARCHRAAIEGRWATTMRFYAGQDLTRSLDFYATFPFP
Mflv_0693|M.gilvum_PYR-GCK -------VIEGSNAERPCYACSGRCCDRYDVR------------------
Mvan_5932|M.vanbaalenii_PYR-1 -------VIVGSVARWPHSACSGRCCD-----------------------
Mb3662|M.bovis_AF2122/97 -HRVLFATASQWVDRLAAAHHSGTLQSELIRLAR-YPLLVVDEVGYIPFE
Rv3638|M.tuberculosis_H37Rv -HRVLFATASQWVDRLAAAHHSGTLQSELIRLAR-YPLLVVDEVGYIPFE
. . .*
MSMEG_2150|M.smegmatis_MC2_155 AEAASALFQVVSQRYLKTSIVITTNRGVGSWGEILGDTTVAAAMLDRLLH
TH_1648|M.thermoresistible__bu AEAASALFQVVSQRYLKTSIVITTNRGVGAWGEILGDTTVAAAMLDRLLH
MAV_1806|M.avium_104 KPTLSKWLRG-DYANLTAVIDLTLSR-LDAG--LFTGTFLEGELTELELQ
Mflv_0693|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5932|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3662|M.bovis_AF2122/97 PEAANLFFQLVSSRYERASLIVTSNKPFGRWGEVFGDDVVAAAMIDRLVH
Rv3638|M.tuberculosis_H37Rv PEAANLFFQLVSSRYERASLIVTSNKPFGRWGEVFGDDVVAAAMIDRLVH
MSMEG_2150|M.smegmatis_MC2_155 RSVVINLDGESYRLRDHQAAAETLRRATTGTRQALH
TH_1648|M.thermoresistible__bu RSVVINLDGESYRLRDHQAAAETLRRTTTGTRQPLH
MAV_1806|M.avium_104 WGRTIGQMPSPYTARNPLVVPYHFNQGP--------
Mflv_0693|M.gilvum_PYR-GCK ------------------------------------
Mvan_5932|M.vanbaalenii_PYR-1 ------------------------------------
Mb3662|M.bovis_AF2122/97 HAEVIALKGDSYRIKDRDLGRVPTVTADDQ------
Rv3638|M.tuberculosis_H37Rv HAEVIALKGDSYRIKDRDLGRVPTVTADDQ------