For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNVVGTRESLALYSDWAYASAVGVLTLALLLLAATFAIGRTRTLERSMAPVGVSLDSASPGVVTEMPRRS RAERMERAGIAVVFVGTVLLLMCIVLRGLATSRAPWGNMYEFINLTCLCGLVAAAVVLRRPQFRALWVFV LIPVLILLAVSGKWLYADAAPVMPALQSYWLPIHVSVVSLGSGVFLVAGVASILFLAKTSTHGELRGDGG NGYVAGIMSRLPDAQVLDRIAYRTTIFAFPTFGFGVIFGAIWAEETWGRFWGWDPKETVSFIAWVAYAAY LHARSTAGWRDRKAAWINVAGFVAMVFNLFFINLVIAGLHSYAGVG
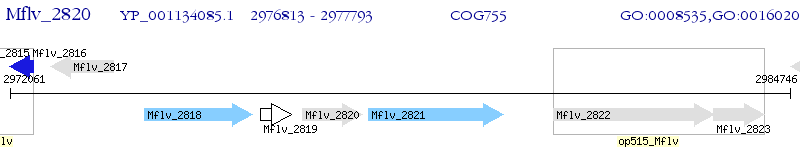
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2820 | - | - | 100% (326) | cytochrome c assembly protein |
| M. gilvum PYR-GCK | Mflv_0049 | - | e-133 | 73.83% (321) | cytochrome c assembly protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0542 | ccsA | 1e-134 | 72.48% (327) | cytochrome C-type biogenesis protein CcsA |
| M. tuberculosis H37Rv | Rv0529 | ccsA | 1e-134 | 72.48% (327) | cytochrome C-type biogenesis protein CcsA |
| M. leprae Br4923 | MLBr_02409 | - | 1e-124 | 66.67% (327) | putative cytochrome C biogenesis protein |
| M. abscessus ATCC 19977 | MAB_3973c | - | 1e-117 | 68.00% (325) | cytochrome c-type biogenesis protein CcsA |
| M. marinum M | MMAR_0875 | ccsB | 1e-132 | 70.43% (328) | cytochrome C-type biogenesis protein, CcsB |
| M. avium 104 | MAV_4616 | ccsB | 1e-136 | 73.17% (328) | cytochrome c-type biogenesis protein CcsB |
| M. smegmatis MC2 155 | MSMEG_0974 | ccsB | 1e-135 | 74.53% (322) | cytochrome c-type biogenesis protein CcsB |
| M. thermoresistible (build 8) | TH_2248 | ccsB | 1e-132 | 73.01% (326) | POSSIBLE CYTOCHROME C-TYPE BIOGENESIS PROTEIN CCSA |
| M. ulcerans Agy99 | MUL_0628 | ccsB | 1e-132 | 70.34% (327) | cytochrome C-type biogenesis protein, CcsB |
| M. vanbaalenii PYR-1 | Mvan_3702 | - | 1e-175 | 94.17% (326) | cytochrome c assembly protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0875|M.marinum_M -MNTANINIDLARYSDWAFTSAVVALVVALLLLAFELAYVRGRKVDEREL
MUL_0628|M.ulcerans_Agy99 -MNTANINIDLARYSDWAFTSAVVALVVALLLLAFELAYVRGRKVDEREL
Mb0542|M.bovis_AF2122/97 -MNTLHVNVGLARYSDWAFTSAVVALVVALLLLAFEFAQVRGRGLAP-LA
Rv0529|M.tuberculosis_H37Rv -MNTLHVNVGLARYSDWAFTSAVVALVVALLLLAFEFAQVRGRGLAP-LA
MLBr_02409|M.leprae_Br4923 -MNTLHVNIGLACYSDWAFTSAVVALVVALLLLAYELAYVGSRRLDNRTR
MAV_4616|M.avium_104 -MNTVHVNIELARYSDWAFTSAVVALVIALLLLAFELAYAGGRRADARAR
MSMEG_0974|M.smegmatis_MC2_155 MMNTEHIDIGLARYSDWAFTSSVLVLVGALLLLAVEMAYSRSRRVESREL
TH_2248|M.thermoresistible__bu --VSTEIDIGLARYSDWAFTTSVLVLMGALLLLAIELAYARSRKVETREL
MAB_3973c|M.abscessus_ATCC_199 -MNSTNIDVNLAKFSDYAFTSAIVVMVGALVLLAIELAYRQSDRAAQREL
Mflv_2820|M.gilvum_PYR-GCK -MNVVGTRESLALYSDWAYASAVGVLTLALLLLAATFAIGRTRTLER-SM
Mvan_3702|M.vanbaalenii_PYR-1 -MNVVGTRESLALYSDWAYASAVGVLTLALVLLSATFAIGWSRTRET-SA
** :**:*::::: .: **:**: :*
MMAR_0875|M.marinum_M VLAG-------SVASDATTPGIVLDGPQRPLDERVGRAGLAVVYLGIGLL
MUL_0628|M.ulcerans_Agy99 VLAG-------SVASDATTPGIVLDGPRRPLDERVGRAGLAVVYLGIGLL
Mb0542|M.bovis_AF2122/97 VPAG-------SVATDSATPGIVADQRHRPFDERVGRGGLAVAYLGIGLL
Rv0529|M.tuberculosis_H37Rv VPAG-------SVATDSATPGIVADQRHRPFDERVGRGGLAVAYLGIGLL
MLBr_02409|M.leprae_Br4923 VVPG-------SVAADSGTPGIVVDSLQRPFGERVGQAGLAVAYLGIGLL
MAV_4616|M.avium_104 VLAG-------AVSADSATPGVVDEVPRRPVDERAGRAGLAVVYLGTGLL
MSMEG_0974|M.smegmatis_MC2_155 VGAG----GPSVVGADSATPGVVVESPKRPFDERVGKTGLALTYVGIGLL
TH_2248|M.thermoresistible__bu AAAGVAASGAVTTSPGAAAPGVVVEAPKRPVDERIGGAGLALAYVGIAAL
MAB_3973c|M.abscessus_ATCC_199 VSSG-----GDVVAAGENRPGLVVPQGRRPLAERVGGAGQALAYLGIGLL
Mflv_2820|M.gilvum_PYR-GCK APVG--------VSLDSASPGVVTEMPRRSRAERMERAGIAVVFVGTVLL
Mvan_3702|M.vanbaalenii_PYR-1 ARVG--------VAPDSASPGVVAVMSRRSRAERMERAGIAVVFVGAVLL
. * .. . **:* :*. ** * *:.::* *
MMAR_0875|M.marinum_M LACIVLRGLATMRVPWGNMYEFINLTCMSGLIAGAVVLRRPQ----YRSL
MUL_0628|M.ulcerans_Agy99 LACIVLRGLATMRVPWGNMYEFINLTCMSGLIAGAVVLRRPQ----YRSL
Mb0542|M.bovis_AF2122/97 LACVVLRGLATQRVPWGNMYEFINLTCLSGLIAGAVVLRRAR----YRPL
Rv0529|M.tuberculosis_H37Rv LACVVLRGLATQRVPWGNMYEFINLTCLSGLIAGAVVLRRAR----YRPL
MLBr_02409|M.leprae_Br4923 LACILLRGLATLRVPWGNMYEFINLTCFSELVAGAIVLRRRQ----YRPL
MAV_4616|M.avium_104 LACLVLRGLATLRVPWGNMYEFINLTCLCGLVAGAIVLRRPQ----YRPL
MSMEG_0974|M.smegmatis_MC2_155 LVCIVLRGVSTSRVPWGNMYEFINLTCFCGLVAAAVVLRRPQ----YRTL
TH_2248|M.thermoresistible__bu LGSIVLRGLATGRAPWGNMYEFVNVTCLCGLVAAAVMLRRAR----YRPL
MAB_3973c|M.abscessus_ATCC_199 LLCIVLRGVATTRVPWGNMYEFINLTCACGLVTAAIVLRPRGNAEGRRAL
Mflv_2820|M.gilvum_PYR-GCK LMCIVLRGLATSRAPWGNMYEFINLTCLCGLVAAAVVLRRPQ----FRAL
Mvan_3702|M.vanbaalenii_PYR-1 LMCIVLRGLATSRAPWGNMYEFINLTCLCGLVAAAVVLRRPQ----FRAL
* .::***::* *.********:*:** . *::.*::** *.*
MMAR_0875|M.marinum_M WVFLLLPVLILLTVSGRWLYTNAAPVMPALQSYWLPIHVSVVSLGSGVFL
MUL_0628|M.ulcerans_Agy99 WVFLLLPVLILLTVSGRWLYTNAAPVMPALQSYWLPIHVSVISLGSGVFL
Mb0542|M.bovis_AF2122/97 WVFLLVPVLILLTVSGRWLYANAAPVMPALQSYWLPIHVSVVSLGSGVFL
Rv0529|M.tuberculosis_H37Rv WVFLLVPVLILLTVSGRWLYANAAPVMPALQSYWLPIHVSVVSLGSGVFL
MLBr_02409|M.leprae_Br4923 WVFLLVSVLILLTISGRWLYANAAPVIPALQSYWLPIHVSVVSLGSGIFL
MAV_4616|M.avium_104 WVFLLLPVLILLTVSGRWLYTNAAPVMPALQSYWLPIHVSVVSLGSGVFL
MSMEG_0974|M.smegmatis_MC2_155 WVFVLVPVLILLTVSGRWLYTNAAPVMPALQSYWLPIHVSVVSLGSGVFL
TH_2248|M.thermoresistible__bu WVFVLVPVLILLTVSGRWLYTDAAPVMPALQSYWLPIHVSVVSLGSGVFL
MAB_3973c|M.abscessus_ATCC_199 WVFVLIPVLILLAVSGRYLYTYAAPVMPALQSYWLPIHVSVVSLGSGVFL
Mflv_2820|M.gilvum_PYR-GCK WVFVLIPVLILLAVSGKWLYADAAPVMPALQSYWLPIHVSVVSLGSGVFL
Mvan_3702|M.vanbaalenii_PYR-1 WVFVLIPVLILLAVSGKWLYADAAPVMPALQSYWLPIHVSVVSLGSGVFL
***:*:.*****::**::**: ****:**************:*****:**
MMAR_0875|M.marinum_M VAGISSILFLLRTSRLGAPDSQPAESTLGRLVQRLPDAQTLDRIAYRTTI
MUL_0628|M.ulcerans_Agy99 VAGISSILFLLRTSRLGAPDSQPAESTLGRLVQRLPDAQTLDRIAYRTTI
Mb0542|M.bovis_AF2122/97 VAGVASILFLVRTSRLG--EPTGEG-ALAGMVRRLPDAQTLDGIAYRTTI
Rv0529|M.tuberculosis_H37Rv VAGVASILFLVRTSRLG--EPTGEG-ALAGMVRRLPDAQTLDGIAYRTTI
MLBr_02409|M.leprae_Br4923 VAGIASILFLLSTSRLVASDPEGAK-ALARLVRRFPDAQTLDRIAYRTTI
MAV_4616|M.avium_104 VAGIASILFLLRTSPLG--DPGNDS-APARLVRRFPDAQTLDRIAYRTTI
MSMEG_0974|M.smegmatis_MC2_155 VAGVASILFLLKMSPLGQPQEGRDS-VWARIIDRVPDAQTLDRIAYRTTI
TH_2248|M.thermoresistible__bu VAGVASILFLVKMSRFG--QVAGDG-LWARIVDRLPDAQTLDRIAYRATI
MAB_3973c|M.abscessus_ATCC_199 VAGVASILFLAKMYFPD-----------NSFVQRLPDAQALDRLAYRTTI
Mflv_2820|M.gilvum_PYR-GCK VAGVASILFLAKTSTHGELRGDGGNGYVAGIMSRLPDAQVLDRIAYRTTI
Mvan_3702|M.vanbaalenii_PYR-1 VAGVASILFLVKTSIHGEQRADGRNGYVAGIMSRLPDAQVLDRIAYRTTI
***::***** :: *.****.** :***:**
MMAR_0875|M.marinum_M FAFPVFGFGVIFGAIWAEQAWGRYWGWDPKETVSFVAWVVYAAYLHARST
MUL_0628|M.ulcerans_Agy99 FAFPVFGFGVIFGAIWAEQAWGRYWGWDPKETVSFVAWVVYAAYLHARST
Mb0542|M.bovis_AF2122/97 FAFPVFGFGVIFGAIWAEEAWGRYWGWDPKETVSFVAWVVYAAYLHARST
Rv0529|M.tuberculosis_H37Rv FAFPVFGFGVIFGAIWAEEAWGRYWGWDPKETVSFVAWVVYAAYLHARST
MLBr_02409|M.leprae_Br4923 FAFPVFGFGVIFGAIWAEQAWGRYWGWDPKETVSFVAWVVYAAYLHARST
MAV_4616|M.avium_104 FAFPVFGFGVIFGAIWAEEAWGRYWGWDPKETVSFVAWVIYAAYLHARST
MSMEG_0974|M.smegmatis_MC2_155 FAFPVFGFGVIFGAIWAEEAWGRYWGWDPKETVSFIAWVVYAAYLHARST
TH_2248|M.thermoresistible__bu FAFPVFGFGVIFGAIWAEEAWGRYWGWDPKETVSFIAWVVYAAYLHARST
MAB_3973c|M.abscessus_ATCC_199 FGFPIFGFGVIFGAIWAEEAWGRFWAWDPKETVSFVAWVIYAAYLHARST
Mflv_2820|M.gilvum_PYR-GCK FAFPTFGFGVIFGAIWAEETWGRFWGWDPKETVSFIAWVAYAAYLHARST
Mvan_3702|M.vanbaalenii_PYR-1 FAFPTFGFGVIFGAIWAEETWGRFWGWDPKETVSFIAWVAYAAYLHARST
*.** *************::***:*.*********:*** **********
MMAR_0875|M.marinum_M AGWRDRKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVG
MUL_0628|M.ulcerans_Agy99 AGWRDRKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVV
Mb0542|M.bovis_AF2122/97 AGWRDRKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVG
Rv0529|M.tuberculosis_H37Rv AGWRDRKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVG
MLBr_02409|M.leprae_Br4923 AGWRDRRAAWINVVGCVAMVFNLFFVNLVTVGLHSYAGVD
MAV_4616|M.avium_104 AGWRDRKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVG
MSMEG_0974|M.smegmatis_MC2_155 AGWRDRKAAWINVVGFVAMVFNLFFINLVTVGLHSYAGVG
TH_2248|M.thermoresistible__bu AGWRDRKAAWINVIGFVAMVFNLFFINLVTVGLHSYAGVG
MAB_3973c|M.abscessus_ATCC_199 AGWRDKKAAWINVAGFVAMVFNLFFVNLVTVGLHSYAGVS
Mflv_2820|M.gilvum_PYR-GCK AGWRDRKAAWINVAGFVAMVFNLFFINLVIAGLHSYAGVG
Mvan_3702|M.vanbaalenii_PYR-1 AGWRDRKAAWINVAGFVAMVFNLFFINLVIAGLHSYAGVG
*****::****** * *********:*** .********