For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVGCPVIGQVVLPTSPPSLWAMLGWLPPPVPILPVLAIVLAVWYLLAVRVIRCRGRQWAWTRTASFLAGC IALAAVTGLAIDGYGYRLFSAFMFQHLTLSILVPPLLVLGAPGRLLLRSTPHRGLGRYVVIAALGGLRCR ASRIVLHPGFTIPVFLLSYYGLYLSDLFDAAASSVAGHVTLQVFFLTSGMLFILPILSTGPLPVRQSNLG RFFDIFVEMPLHVFIGVILMMAPRTLTETFAQPPPDWNVDPVADQGVAGALAWSYGEPVALLTTVIFAIR WRRDEESESKKREADVERDDAELAAYNAFLRGLHQQRRPPTDVPSTANEHD
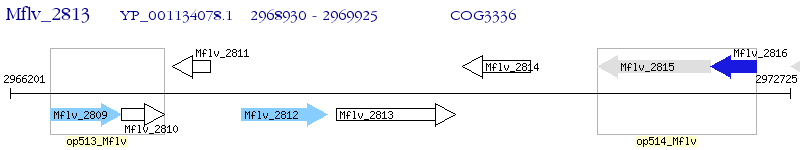
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2813 | - | - | 100% (331) | hypothetical protein Mflv_2813 |
| M. gilvum PYR-GCK | Mflv_2566 | - | 1e-48 | 38.44% (294) | copper resistance D domain-containing protein |
| M. gilvum PYR-GCK | Mflv_2826 | - | 3e-24 | 30.45% (266) | copper resistance D domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0105 | - | 6e-39 | 33.90% (292) | integral membrane protein |
| M. tuberculosis H37Rv | Rv0102 | - | 6e-39 | 33.90% (292) | integral membrane protein |
| M. leprae Br4923 | MLBr_01998 | - | 6e-34 | 31.51% (292) | hypothetical protein MLBr_01998 |
| M. abscessus ATCC 19977 | MAB_4494c | - | 1e-122 | 66.12% (307) | hypothetical protein MAB_4494c |
| M. marinum M | MMAR_0267 | - | 8e-35 | 32.42% (293) | integral membrane protein |
| M. avium 104 | MAV_1692 | - | 7e-47 | 37.97% (295) | ABC-type transporter, permease components |
| M. smegmatis MC2 155 | MSMEG_4702 | - | 2e-46 | 37.11% (291) | ABC-type transporter, permease components |
| M. thermoresistible (build 8) | TH_1033 | - | 8e-34 | 37.99% (229) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4847 | - | 1e-34 | 31.89% (301) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_3709 | - | 0.0 | 97.24% (326) | hypothetical protein Mvan_3709 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0267|M.marinum_M MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
MUL_4847|M.ulcerans_Agy99 MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
Mb0105|M.bovis_AF2122/97 MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
Rv0102|M.tuberculosis_H37Rv MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
MLBr_01998|M.leprae_Br4923 MQTDEPTGTATSAIRIPPLISLRTTLMVAGLLGNVVAVAAYGLFSRTERY
MAV_1692|M.avium_104 MTVAPPAGTASTEAVPQHRTQVWPVLAGVAVLAG-CTAAGIGTLSLASAL
MSMEG_4702|M.smegmatis_MC2_155 --------------MASDVRAVWPVLVGVGLLAG-AVAAGIGALALADAL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M ATVG---------NPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
MUL_4847|M.ulcerans_Agy99 ATVGNPYPDATVGNPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
Mb0105|M.bovis_AF2122/97 AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
Rv0102|M.tuberculosis_H37Rv AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
MLBr_01998|M.leprae_Br4923 GAVG---------NPDPGSFIGVAEPVGYFGASLSSALCLGALIYFVMMV
MAV_1692|M.avium_104 TATG---------LPDPGPVTTLGLPFLRAAGEIAAVLAVGSFLFAAFLV
MSMEG_4702|M.smegmatis_MC2_155 TATG---------LPNPGPVTTYGLPFVRAAGEIAAVIAVGSFLFAAFLV
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
MUL_4847|M.ulcerans_Agy99 RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
Mb0105|M.bovis_AF2122/97 KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
Rv0102|M.tuberculosis_H37Rv KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
MLBr_01998|M.leprae_Br4923 WPQADGLIDAQAFRIHLAEERVSIGWLAFALTMVVVQAANDSGVTPAELR
MAV_1692|M.avium_104 PPQPSGVLDADGYRALRLGTVASGVWAVCAALLVPLTLSDVSGHPVAD-L
MSMEG_4702|M.smegmatis_MC2_155 APQENGVLDVAGYRALRLGSAASAVWTVCAAMLVPLTISDVSGQPVLEHL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M ASGAVRDAIATSEIARAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
MUL_4847|M.ulcerans_Agy99 ASGAVRDAIATSEIAHAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
Mb0105|M.bovis_AF2122/97 ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
Rv0102|M.tuberculosis_H37Rv ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
MLBr_01998|M.leprae_Br4923 TGAALFNAITTSETSRAWIVVTICALLVTLTLRVIIRYSDHFVLLIPAVI
MAV_1692|M.avium_104 RPAQMWSLAGLITTASAWRWTAILAAAITLASLPVLRWSWAPVLVAASLT
MSMEG_4702|M.smegmatis_MC2_155 SPVDVWSATSLVDIAGAWRWTAILAAAVTIASLPVLRWGWTPVLFAGSLV
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
MUL_4847|M.ulcerans_Agy99 AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
Mb0105|M.bovis_AF2122/97 AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
Rv0102|M.tuberculosis_H37Rv AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
MLBr_01998|M.leprae_Br4923 GVIATAVTGNAGQGPDHDYATSAVIVFALAIAALSG---LKITAALTEVT
MAV_1692|M.avium_104 TLIPLGLTGHSSAGGSHDLATNGLLIHLVAAALWSGGLLALLAYALRGGQ
MSMEG_4702|M.smegmatis_MC2_155 TLVPLALTGHSSSGGAHDLATNSLFIHLVAGAVWAGGLLALLAHAMRGRA
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M P--------TRTVQLMQVACGALAVSYGAVLLYLFVPG-WDFGSEFAR--
MUL_4847|M.ulcerans_Agy99 P--------TRAVQLMQVACGALAVSYGTVLLYLFVPG-WDFGSEFAR--
Mb0105|M.bovis_AF2122/97 P--------SRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFAR--
Rv0102|M.tuberculosis_H37Rv P--------SRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFAR--
MLBr_01998|M.leprae_Br4923 P--------SRAVLIVQVVCGVLALAYGAELVYLLSPGAAMIDSDFGR--
MAV_1692|M.avium_104 G-GGHLGLATRRFSAIALWCWVAMALSGLVNAAVRVQPSDLLATGYGRLV
MSMEG_4702|M.smegmatis_MC2_155 GEGAATALAARRFSALALWCFIAMGVSGVINALVRMSPADLFRTSYGWLI
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M IGLLAGLVLVLVWVFDCWQLFVRPPRSTR---GATAAALAMMAVLAAIG-
MUL_4847|M.ulcerans_Agy99 IGLLAGLLLVLVWVFDCWQLFVRPPRSTR---GATAAALAMMAVLAAIG-
Mb0105|M.bovis_AF2122/97 LGLLAGVILTSVWLFDCWRLLVRPPHAGRRRGGGSGAALAMMAAMASIA-
Rv0102|M.tuberculosis_H37Rv LGLLAGVILTSVWLFDCWRLLVRPPHAGRRRGGGSGAALAMMAAMASIA-
MLBr_01998|M.leprae_Br4923 LGLVAGVLVTLVCFSDCWALVMGRPHSDHR---TRLTALAMMAVTAATA-
MAV_1692|M.avium_104 LAKAAALCLLGGVGWRQRRVNVAALQAVSTLARARRALLRLTLIEAALFG
MSMEG_4702|M.smegmatis_MC2_155 VGKIVALVVLGALGFLQRRSAVSALQADPT---ARGPLIRLALTEGAIFG
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3709|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4494c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0267|M.marinum_M -------AMAVQTAPRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
MUL_4847|M.ulcerans_Agy99 -------AMAVQTAPRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
Mb0105|M.bovis_AF2122/97 -------AMAVMTAPRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
Rv0102|M.tuberculosis_H37Rv -------AMAVMTAPRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
MLBr_01998|M.leprae_Br4923 -------AMSVQTAPRFLTHRFTAWDVFLGYELPQPPNIVRFFTVWRFDS
MAV_1692|M.avium_104 LTFGIAVGLGRTAPPPPPARLPSIAEAEIGYDFDGPPTLTRILFDWRFDL
MSMEG_4702|M.smegmatis_MC2_155 VTFGIAVGLGRT-PPPPIKRIPSAAEVAIGYDFAGPPTVARVLFDWRFDL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK ------MVGCPVIGQVVLPTSPPSLWAMLGWLPPPVP-------------
Mvan_3709|M.vanbaalenii_PYR-1 -----------MIGQVVLPTSPPSLWAMLGWLPPPVP-------------
MAB_4494c|M.abscessus_ATCC_199 ------MNEDSSRGLTSIPDEPPSAWSLLEWNPPTIP-------------
MMAR_0267|M.marinum_M LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
MUL_4847|M.ulcerans_Agy99 LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
Mb0105|M.bovis_AF2122/97 LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
Rv0102|M.tuberculosis_H37Rv LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
MLBr_01998|M.leprae_Br4923 LIGATAIVLGIGYVAGFVRVRRAGNTWPVGRLVAWLTGCVALVFTSSSGV
MAV_1692|M.avium_104 IFGTAAIVLAGLYVAGVVRLRRRGDRWQPGCTSSWLLGCLVLLFVTSSGV
MSMEG_4702|M.smegmatis_MC2_155 LFGTAAIVGALVYLAALYRLRHRGDAWPVGRTVAWLLGCATLLFTTSSGL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Mflv_2813|M.gilvum_PYR-GCK ILPVLAIVLAVWYLLAVRVIRCRGRQWAWTRTASFLAGCIALAAVTGLAI
Mvan_3709|M.vanbaalenii_PYR-1 ILPVLAIVLAVWYLLAVRVVRSRGRQWAWTRTASFLAGCIALAAVTGLAI
MAB_4494c|M.abscessus_ATCC_199 VLPVIAVLLAGWYVLSVLRLRRMGRSWPWWSTVCFLSGCAILGAVMGLSI
MMAR_0267|M.marinum_M RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAGEGQPPGP
MUL_4847|M.ulcerans_Agy99 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAVEGQPPGP
Mb0105|M.bovis_AF2122/97 RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
Rv0102|M.tuberculosis_H37Rv RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
MLBr_01998|M.leprae_Br4923 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALSAAGDGQQPGP
MAV_1692|M.avium_104 GRYMPAMFSMHMVVHMCLSMLVPILLALGAPVTLALRALPAAGRGDPPGP
MSMEG_4702|M.smegmatis_MC2_155 GRYMPAMFSMHMIAHMLLSMLVPILLVLGAPVTMALRALPAAGRGQPPGP
TH_1033|M.thermoresistible__bu ---------------------VPVLLALGGPVTLALRALPVAGRGAPPGP
Mflv_2813|M.gilvum_PYR-GCK DGYGYRLFSAFMFQHLTLSILVPPLLVLGAPGRLLLRSTPHRGLGR--YV
Mvan_3709|M.vanbaalenii_PYR-1 DGYGYRLFSAFMFQHLTLSILVPPLLVLGAPGRLLLRSTPHRGLGR--YV
MAB_4494c|M.abscessus_ATCC_199 ERYGFRLFSAFMFQQLTLSILVPPLLVLGSPGRLLLRSTPHHGSGR--WV
:* **.**.* : ** . *
MMAR_0267|M.marinum_M REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
MUL_4847|M.ulcerans_Agy99 REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
Mb0105|M.bovis_AF2122/97 REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
Rv0102|M.tuberculosis_H37Rv REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
MLBr_01998|M.leprae_Br4923 REWLTWLMHSSVTAFLSHPVTNFILFVGSPYIVYFTPLFDTLVRYHWGHE
MAV_1692|M.avium_104 REWLLAALHSRFSRLLTNPVVATVLFVAGFYGLYLSNLFDTTASSHAGHL
MSMEG_4702|M.smegmatis_MC2_155 REWLLAALHSKISQFFTNPVIATVMFVVGFYGLYFGGIFDAAVSNHAAHV
TH_1033|M.thermoresistible__bu REWLLAALHSPPSRFFTHPLVATVMFVVGFYGLYFGGLFDVAAKEHAAHV
Mflv_2813|M.gilvum_PYR-GCK VIAALGGLRCRASRIVLHPGFTIPVFLLSYYGLYLSDLFDAAASSVAGHV
Mvan_3709|M.vanbaalenii_PYR-1 MISALGGLRCRASRIVLHPGFTIPVFLLSYYGLYLSDLFDAAAASVAGHV
MAB_4494c|M.abscessus_ATCC_199 LAAALAGLRSRAGALLLHPAVTIPLFLFSYYGLYLSQLFDTIAATWLGHT
::. :. :* :*: . * :*: :**. . .*
MMAR_0267|M.marinum_M FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MUL_4847|M.ulcerans_Agy99 FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
Mb0105|M.bovis_AF2122/97 FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
Rv0102|M.tuberculosis_H37Rv FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MLBr_01998|M.leprae_Br4923 FMAIHFLLVGYIFYWAIIGIDPGPRRLPYLARIGLLFAVMPFHAFFGIAL
MAV_1692|M.avium_104 LMNLHFLLSGYLFYWVVIGVDPTPRPIPPLAKLAVVFASLPLHAFFGVVL
MSMEG_4702|M.smegmatis_MC2_155 MMNVHFLLSGYLFYWVVIGIDPTPRPIPHLGKIGMVFASLPLHAFFGVVM
TH_1033|M.thermoresistible__bu AMNIHFLLSGYLFYWVAIGVDPPPRQVPHLAKVGMVFASLPLHAFFGVAL
Mflv_2813|M.gilvum_PYR-GCK TLQVFFLTSGMLFILPILSTGPLPVRQSNLGRFFDIFVEMPLHVFIGVIL
Mvan_3709|M.vanbaalenii_PYR-1 TLQVFFLASGLLFILPILSTGPLPVRQSNLGRFFDIFVEMPLHVFIGVIL
MAB_4494c|M.abscessus_ATCC_199 TLEVFFLATGLLFVIPILSTDPLPIRQTNLGRMFDIFVEMPLHVFIGVVL
: :.** * :* :. .* * . .:. :*. :*:*.*:*: :
MMAR_0267|M.marinum_M MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
MUL_4847|M.ulcerans_Agy99 MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
Mb0105|M.bovis_AF2122/97 MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
Rv0102|M.tuberculosis_H37Rv MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
MLBr_01998|M.leprae_Br4923 MTMTSAISAEFYRSVNLPWLASIDADQHIGGAIAWSATELPVIIVIVALM
MAV_1692|M.avium_104 MGTRKVLGADYYRSLGFSWHTDLLGDQRLGGGIAWSAGEFPLVIVMLALL
MSMEG_4702|M.smegmatis_MC2_155 MGMKTVLGEPFYRSLQLSWHTDLLGDQKLGGGVAWAAGEVPLVIVMLALL
TH_1033|M.thermoresistible__bu MGMKNILAERFYRSLLLDWHTDLAADQRLGGAIAWAAGEVPLVLVMLALL
Mflv_2813|M.gilvum_PYR-GCK MMAPRTLTETFAQPP-PDWNVDPVADQGVAGALAWSYGEPVALLTTVIFA
Mvan_3709|M.vanbaalenii_PYR-1 MMAPRTLTETFSQPP-PDWNVDPVADQGVAGALAWSYGEPVALLTTVIFA
MAB_4494c|M.abscessus_ATCC_199 MMAPRPLIGIFANPP-AQWSVDPVRDQAVAGALAWSYGEPIALATTLIFA
* : : .. * . ** :.*.:**. * : . : :
MMAR_0267|M.marinum_M AQWARQDRQVAARQDRHADSSYADDDLEAYNAMLRELARMRE--------
MUL_4847|M.ulcerans_Agy99 AQWARQDRQVAARKDRHADSSYADDDLEAYNAMLRELARMRE--------
Mb0105|M.bovis_AF2122/97 TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
Rv0102|M.tuberculosis_H37Rv TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
MLBr_01998|M.leprae_Br4923 AQWARQDHRVAVREDRHADSTYADDDLNAYNAMLRELSRMRR--------
MAV_1692|M.avium_104 VQWARSDRRTAKRLDRAADRD-DDAELAAYNAMLAQLAGRDKPGEGSATG
MSMEG_4702|M.smegmatis_MC2_155 IQWRRSDDRTAKRLDRAADRD-DDADLAAYNRMLAHLARRDNPPQ-----
TH_1033|M.thermoresistible__bu IQWSRSDQRAARRMDRAADRD-HDAELAAYNAMLAELTRREQQRTRTER-
Mflv_2813|M.gilvum_PYR-GCK IRWRRDEE--SESKKREADVERDDAELAAYNAFLRGLHQQRRPPTDVPST
Mvan_3709|M.vanbaalenii_PYR-1 IRWRRDEE--SESKKREADVERDDAELAAYNAFLRGLHQQRRPPTDLPST
MAB_4494c|M.abscessus_ATCC_199 VRWRREEQ--AETEEREADNARQEDELASYNRFLRQLHG---DTTDLNP-
:* *.: : .* ** : :* :** :* *
MMAR_0267|M.marinum_M -----
MUL_4847|M.ulcerans_Agy99 -----
Mb0105|M.bovis_AF2122/97 -----
Rv0102|M.tuberculosis_H37Rv -----
MLBr_01998|M.leprae_Br4923 -----
MAV_1692|M.avium_104 QA---
MSMEG_4702|M.smegmatis_MC2_155 -----
TH_1033|M.thermoresistible__bu -----
Mflv_2813|M.gilvum_PYR-GCK ANEHD
Mvan_3709|M.vanbaalenii_PYR-1 ANEHD
MAB_4494c|M.abscessus_ATCC_199 -----