For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRYAEAGNPYPGAFVSVAEPVGF FAASLAGALCLGALIHVVMTAKPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARL LASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVLAVVATAVTGNPGQGPDHDY ATSAAIVFAVAFATLTGLKIAAALAGTTPSRAVLVTQVTCGALALAYGAMLLYLFIPGWAVDSDFARLGL LAGVILTSVWLFDCWRLLVRPPHAGRRRGGGSGAALAMMAAMASIAAMAVMTAPRFLTHAFTAWDVFLGY ELPQPPTIARVLTVWRFDSLIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGVR AYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGAREWLTWLLHSRVTTFLSHPIT AFVLFVASPYIVYFTPLFDTFVRYHWGHEFMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMP FHAFFGIALMTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALVTQWARQDRRVA SREDRHADSDYADDELEAYNAMLRELSRMRR
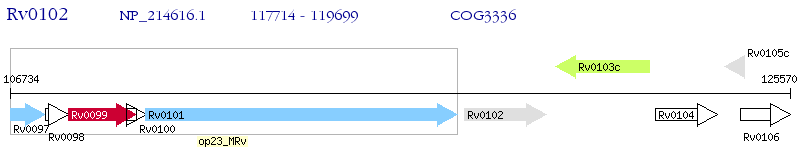
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0102 | - | - | 100% (661) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0105 | - | 0.0 | 100.00% (661) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2566 | - | 1e-110 | 37.02% (678) | copper resistance D domain-containing protein |
| M. leprae Br4923 | MLBr_01998 | - | 0.0 | 70.24% (662) | hypothetical protein MLBr_01998 |
| M. abscessus ATCC 19977 | MAB_1557 | - | 1e-108 | 35.79% (665) | copper resistance D precursor |
| M. marinum M | MMAR_0267 | - | 0.0 | 74.39% (660) | integral membrane protein |
| M. avium 104 | MAV_1692 | - | 1e-108 | 36.98% (676) | ABC-type transporter, permease components |
| M. smegmatis MC2 155 | MSMEG_4702 | - | 1e-115 | 37.07% (661) | ABC-type transporter, permease components |
| M. thermoresistible (build 8) | TH_1033 | - | 2e-66 | 54.59% (218) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4847 | - | 0.0 | 73.09% (669) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4073 | - | 1e-107 | 36.42% (648) | copper resistance D domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2566|M.gilvum_PYR-GCK -----MTSTAPHVRSGVRRAPAWAVLYRVAALAG-VTAVALAALSLADAL
Mvan_4073|M.vanbaalenii_PYR-1 -----MTSVAPVAPGGVRRAPVWAVLYGVAVLAG-VTAAALSALSLADAL
MSMEG_4702|M.smegmatis_MC2_155 ------------MASDVR--AVWPVLVGVGLLAG-AVAAGIGALALADAL
MAV_1692|M.avium_104 MTVAPPAGTASTEAVPQHRTQVWPVLAGVAVLAG-CTAAGIGTLSLASAL
MAB_1557|M.abscessus_ATCC_1997 --------MTTAPTARKTDSPIWTIVVGLAVLAG-LVAATIGALSLAEAL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
Mb0105|M.bovis_AF2122/97 MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
MMAR_0267|M.marinum_M MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
MUL_4847|M.ulcerans_Agy99 MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
MLBr_01998|M.leprae_Br4923 MQTDEPTGTATSAIRIPPLISLRTTLMVAGLLGNVVAVAAYGLFSRTERY
Mflv_2566|M.gilvum_PYR-GCK TATG---------LPDPGPVTSYGLPFVRAAGEIAAVVAVGGFLFAAFMV
Mvan_4073|M.vanbaalenii_PYR-1 TATG---------LPDPGPVTSYGLPFVKAVGEIAAVTATGGFLFAAFMV
MSMEG_4702|M.smegmatis_MC2_155 TATG---------LPNPGPVTTYGLPFVRAAGEIAAVIAVGSFLFAAFLV
MAV_1692|M.avium_104 TATG---------LPDPGPVTTLGLPFLRAAGEIAAVLAVGSFLFAAFLV
MAB_1557|M.abscessus_ATCC_1997 LATG---------LPNPGPVTSYGLPFVRAAAEIAAVVAVGAFLLAAFFV
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
Mb0105|M.bovis_AF2122/97 AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
MMAR_0267|M.marinum_M ATVG---------NPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
MUL_4847|M.ulcerans_Agy99 ATVGNPYPDATVGNPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
MLBr_01998|M.leprae_Br4923 GAVG---------NPDPGSFIGVAEPVGYFGASLSSALCLGALIYFVMMV
Mflv_2566|M.gilvum_PYR-GCK PPQHNQVLDAAGYRALRMGTAASGIWTVCAVLMVPLTVSDVSGQPLSTRM
Mvan_4073|M.vanbaalenii_PYR-1 PPQQNQVLDAAGYRALRTGTAAAGAWAVCAVLMVPLTVSDVSGQPLATRL
MSMEG_4702|M.smegmatis_MC2_155 APQENGVLDVAGYRALRLGSAASAVWTVCAAMLVPLTISDVSGQPVLEHL
MAV_1692|M.avium_104 PPQPSGVLDADGYRALRLGTVASGVWAVCAALLVPLTLSDVSGHPVAD-L
MAB_1557|M.abscessus_ATCC_1997 PPQTSGVLDVDGYRALRLGTAAATVLAVLSAVMVPLSVSDVSGQPLSD-F
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
Mb0105|M.bovis_AF2122/97 KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
MMAR_0267|M.marinum_M RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
MUL_4847|M.ulcerans_Agy99 RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
MLBr_01998|M.leprae_Br4923 WPQADGLIDAQAFRIHLAEERVSIGWLAFALTMVVVQAANDSGVTPAELR
Mflv_2566|M.gilvum_PYR-GCK SPADIWSVADLIDIAGAWRWTALFAALVTVVSIPVLRWSWTPVLLASSLI
Mvan_4073|M.vanbaalenii_PYR-1 SPADIWSVADLIEIAGAWRWTALLAVLVTVVSIPVLRWSWTPVLFAGSLL
MSMEG_4702|M.smegmatis_MC2_155 SPVDVWSATSLVDIAGAWRWTAILAAAVTIASLPVLRWGWTPVLFAGSLV
MAV_1692|M.avium_104 RPAQMWSLAGLITTASAWRWTAILAAAITLASLPVLRWSWAPVLVAASLT
MAB_1557|M.abscessus_ATCC_1997 GPGELWTLASEIETVNAWRWMAFIAAGVAITSRVVLRWSWTPLLVLGSVA
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
Mb0105|M.bovis_AF2122/97 ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
MMAR_0267|M.marinum_M ASGAVRDAIATSEIARAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
MUL_4847|M.ulcerans_Agy99 ASGAVRDAIATSEIAHAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
MLBr_01998|M.leprae_Br4923 TGAALFNAITTSETSRAWIVVTICALLVTLTLRVIIRYSDHFVLLIPAVI
Mflv_2566|M.gilvum_PYR-GCK TLLPIALSGHSSSGGAHDIATNSLTAHMLAGALWAGGLLALLVHALRGSD
Mvan_4073|M.vanbaalenii_PYR-1 TLLPLALSGHSSSGGSHDIATNSLLIHLVAGAIWAGGLLALLVHTLRGGG
MSMEG_4702|M.smegmatis_MC2_155 TLVPLALTGHSSSGGAHDLATNSLFIHLVAGAVWAGGLLALLAHAMRGRA
MAV_1692|M.avium_104 TLIPLGLTGHSSAGGSHDLATNGLLIHLVAAALWSGGLLALLAYALRGGQ
MAB_1557|M.abscessus_ATCC_1997 TLMPLALVGHSATGGAHDLGTNSLIIHLVSAALWAGGLVSLLIHALRGGG
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
Mb0105|M.bovis_AF2122/97 AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
MMAR_0267|M.marinum_M AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
MUL_4847|M.ulcerans_Agy99 AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
MLBr_01998|M.leprae_Br4923 GVIATAVTGNAGQGPDHDYATSAVIVFALAIAALSG---LKITAALTEVT
Mflv_2566|M.gilvum_PYR-GCK DGDDHAALAARRFSAVALWCFVAMALSGTINALVRVRPADLVTTTYGTLL
Mvan_4073|M.vanbaalenii_PYR-1 ----HGPLAARRFSAVALWCFVAMAFSGVINALVRIRPADLVSTQYGLLI
MSMEG_4702|M.smegmatis_MC2_155 GEGAATALAARRFSALALWCFIAMGVSGVINALVRMSPADLFRTSYGWLI
MAV_1692|M.avium_104 GGG-HLGLATRRFSAIALWCWVAMALSGLVNAAVRVQPSDLLATGYGRLV
MAB_1557|M.abscessus_ATCC_1997 ----HLDVAARRFSMLALWCYVAIGLSGIINALIRVQLSDLFTTRYGWLL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv --------PSRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFARLG
Mb0105|M.bovis_AF2122/97 --------PSRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFARLG
MMAR_0267|M.marinum_M --------PTRTVQLMQVACGALAVSYGAVLLYLFVPG-WDFGSEFARIG
MUL_4847|M.ulcerans_Agy99 --------PTRAVQLMQVACGALAVSYGTVLLYLFVPG-WDFGSEFARIG
MLBr_01998|M.leprae_Br4923 --------PSRAVLIVQVVCGVLALAYGAELVYLLSPGAAMIDSDFGRLG
Mflv_2566|M.gilvum_PYR-GCK LAKIAALVALGVLGWQQRRRGVAALQAD---PRARGPLIRLALVEAAVFG
Mvan_4073|M.vanbaalenii_PYR-1 LAKLVALVTLGGLGWQQRRRAVAALQQD---PDARQPLIRLALVEAVVFG
MSMEG_4702|M.smegmatis_MC2_155 VGKIVALVVLGALGFLQRRSAVSALQAD---PTARGPLIRLALTEGAIFG
MAV_1692|M.avium_104 LAKAAALCLLGGVGWRQRRVNVAALQAVSTLARARRALLRLTLIEAALFG
MAB_1557|M.abscessus_ATCC_1997 MGKATALLLLGVLGYLQRRSAITALDED---PENRQPLIRLAGVEAIIFA
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv LLAGVILTSVWLFDCWRLLVRP---------PHAGRRRGGGSGAALAMMA
Mb0105|M.bovis_AF2122/97 LLAGVILTSVWLFDCWRLLVRP---------PHAGRRRGGGSGAALAMMA
MMAR_0267|M.marinum_M LLAGLVLVLVWVFDCWQLFVRP---------PRSTR---GATAAALAMMA
MUL_4847|M.ulcerans_Agy99 LLAGLLLVLVWVFDCWQLFVRP---------PRSTR---GATAAALAMMA
MLBr_01998|M.leprae_Br4923 LVAGVLVTLVCFSDCWALVMGR---------PHSDHR---TRLTALAMMA
Mflv_2566|M.gilvum_PYR-GCK LTFGVAVALGRTPPPPPLNPNLSIPAVEIGYDLDGPPTLGRLLFDWRFDL
Mvan_4073|M.vanbaalenii_PYR-1 LTFGVAVALGRTPPPPPTNTNLSIPAVEIGYDLAGPPTVGKVLFDWRFDL
MSMEG_4702|M.smegmatis_MC2_155 VTFGIAVGLGRTPPPP-IKRIPSAAEVAIGYDFAGPPTVARVLFDWRFDL
MAV_1692|M.avium_104 LTFGIAVGLGRTAPPPPPARLPSIAEAEIGYDFDGPPTLTRILFDWRFDL
MAB_1557|M.abscessus_ATCC_1997 VTFGIAVGLGRTPPPPPVNLNPSPVEVAIGYNLDGPPTPGRLMFDWRFDL
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv AMASIAAMAVMT-APRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
Mb0105|M.bovis_AF2122/97 AMASIAAMAVMT-APRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
MMAR_0267|M.marinum_M VLAAIGAMAVQT-APRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
MUL_4847|M.ulcerans_Agy99 VLAAIGAMAVQT-APRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
MLBr_01998|M.leprae_Br4923 VTAATAAMSVQT-APRFLTHRFTAWDVFLGYELPQPPNIVRFFTVWRFDS
Mflv_2566|M.gilvum_PYR-GCK VLGTAAIVLALVYLAGVRRLRRRGDAWPTGRVVAWLLGCLVLLLATSSGF
Mvan_4073|M.vanbaalenii_PYR-1 IFGTAAIVFALVYLAGVRRLRRRGDAWPIGRVVAWLLGCLVLLLATSSGI
MSMEG_4702|M.smegmatis_MC2_155 LFGTAAIVGALVYLAALYRLRHRGDAWPVGRTVAWLLGCATLLFTTSSGL
MAV_1692|M.avium_104 IFGTAAIVLAGLYVAGVVRLRRRGDRWQPGCTSSWLLGCLVLLFVTSSGV
MAB_1557|M.abscessus_ATCC_1997 IFGTAAIVFAIAYVVGVRRLKQRGDEWPIGRTISWLFGCGFLLIATSSGV
TH_1033|M.thermoresistible__bu --------------------------------------------------
Rv0102|M.tuberculosis_H37Rv LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
Mb0105|M.bovis_AF2122/97 LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
MMAR_0267|M.marinum_M LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
MUL_4847|M.ulcerans_Agy99 LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
MLBr_01998|M.leprae_Br4923 LIGATAIVLGIGYVAGFVRVRRAGNTWPVGRLVAWLTGCVALVFTSSSGV
Mflv_2566|M.gilvum_PYR-GCK GRYMPAMFSIHMIAHMMLSMLAPILLVLGAPVTLALRALPVAGRTGAPGP
Mvan_4073|M.vanbaalenii_PYR-1 GRYMPAMFSMHMIAHMLLSMLAPILLVLGAPVTLALRALPAAGRDGPPGP
MSMEG_4702|M.smegmatis_MC2_155 GRYMPAMFSMHMIAHMLLSMLVPILLVLGAPVTMALRALPAAGRGQPPGP
MAV_1692|M.avium_104 GRYMPAMFSMHMVVHMCLSMLVPILLALGAPVTLALRALPAAGRGDPPGP
MAB_1557|M.abscessus_ATCC_1997 GRYMPAMFSMHMVAHMMLSMLVPVLLVLGGPVTLLLRVLPAAGKSDPPGL
TH_1033|M.thermoresistible__bu ---------------------VPVLLALGGPVTLALRALPVAGRGAPPGP
Rv0102|M.tuberculosis_H37Rv RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
Mb0105|M.bovis_AF2122/97 RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
MMAR_0267|M.marinum_M RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAGEGQPPGP
MUL_4847|M.ulcerans_Agy99 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAVEGQPPGP
MLBr_01998|M.leprae_Br4923 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALSAAGDGQQPGP
*:**.**.***: **.*..: **
Mflv_2566|M.gilvum_PYR-GCK REWLLAALHSRVSQFLTNPFVATGLFVVGFYGLYFSGLFDAAAGSHAAHV
Mvan_4073|M.vanbaalenii_PYR-1 REWLLAALHSRYSQVVTNPIVATALFVVGFYGLYFSGLFEAAAGSHFAHL
MSMEG_4702|M.smegmatis_MC2_155 REWLLAALHSKISQFFTNPVIATVMFVVGFYGLYFGGIFDAAVSNHAAHV
MAV_1692|M.avium_104 REWLLAALHSRFSRLLTNPVVATVLFVAGFYGLYLSNLFDTTASSHAGHL
MAB_1557|M.abscessus_ATCC_1997 REWVQLALHSKFSRFLTHPLVATSLFIGGFYGLYLSGLYDAAVDVHAAHL
TH_1033|M.thermoresistible__bu REWLLAALHSPPSRFFTHPLVATVMFVVGFYGLYFGGLFDVAAKEHAAHV
Rv0102|M.tuberculosis_H37Rv REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
Mb0105|M.bovis_AF2122/97 REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
MMAR_0267|M.marinum_M REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
MUL_4847|M.ulcerans_Agy99 REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
MLBr_01998|M.leprae_Br4923 REWLTWLMHSSVTAFLSHPVTNFILFVGSPYIVYFTPLFDTLVRYHWGHE
***: :** : ..::*. :*: . * :*: :::. . * .*
Mflv_2566|M.gilvum_PYR-GCK AMNVHFLLSGYLFYWVVIGVDPTPRPIPHLAKLGMVFASLPLHAFFGVVL
Mvan_4073|M.vanbaalenii_PYR-1 AMNLHFLVSGYLFYWVVIGIDPTPRPIPHLAKLAVVFASLPLHAFFGVVL
MSMEG_4702|M.smegmatis_MC2_155 MMNVHFLLSGYLFYWVVIGIDPTPRPIPHLGKIGMVFASLPLHAFFGVVM
MAV_1692|M.avium_104 LMNLHFLLSGYLFYWVVIGVDPTPRPIPPLAKLAVVFASLPLHAFFGVVL
MAB_1557|M.abscessus_ATCC_1997 AMNLHFLLSGYLFYWVVIGIDPSPRRLPPVAKLGIVLVSLPLHAFFGVIL
TH_1033|M.thermoresistible__bu AMNIHFLLSGYLFYWVAIGVDPPPRQVPHLAKVGMVFASLPLHAFFGVAL
Rv0102|M.tuberculosis_H37Rv FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
Mb0105|M.bovis_AF2122/97 FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MMAR_0267|M.marinum_M FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MUL_4847|M.ulcerans_Agy99 FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MLBr_01998|M.leprae_Br4923 FMAIHFLLVGYIFYWAIIGIDPGPRRLPYLARIGLLFAVMPFHAFFGIAL
* :***: **:***. **:** ** :* .::.:::. :*:*****: :
Mflv_2566|M.gilvum_PYR-GCK MGTQSVLAESFYRSLQLPWHTDLLADQRVGGGIAWAAGEVPLVVVLIALF
Mvan_4073|M.vanbaalenii_PYR-1 MGTQSVLAESFYRSLQLSWHTDLMGDQKLGGGIAWAAGEIPLVVVLIALF
MSMEG_4702|M.smegmatis_MC2_155 MGMKTVLGEPFYRSLQLSWHTDLLGDQKLGGGVAWAAGEVPLVIVMLALL
MAV_1692|M.avium_104 MGTRKVLGADYYRSLGFSWHTDLLGDQRLGGGIAWSAGEFPLVIVMLALL
MAB_1557|M.abscessus_ATCC_1997 MGTKSILGEKFYSNLALPWRIDLAADQHMGGAITWATGELPLMVVMIALV
TH_1033|M.thermoresistible__bu MGMKNILAERFYRSLLLDWHTDLAADQRLGGAIAWAAGEVPLVLVMLALL
Rv0102|M.tuberculosis_H37Rv MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
Mb0105|M.bovis_AF2122/97 MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
MMAR_0267|M.marinum_M MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
MUL_4847|M.ulcerans_Agy99 MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
MLBr_01998|M.leprae_Br4923 MTMTSAISAEFYRSVNLPWLASIDADQHIGGAIAWSATELPVIIVIVALM
* . :. :* .: : * .: .**::**.::*. *.*:::*::**.
Mflv_2566|M.gilvum_PYR-GCK IQWRRTDDRTARRLDRAADRD-DDADLAAYNNMLAELARRERERG-----
Mvan_4073|M.vanbaalenii_PYR-1 VQWRRTDDRTAKRLDRAADRD-DDADLAAYNAMLAELARRERERS-----
MSMEG_4702|M.smegmatis_MC2_155 IQWRRSDDRTAKRLDRAADRD-DDADLAAYNRMLAHLARRDNPPQ-----
MAV_1692|M.avium_104 VQWARSDRRTAKRLDRAADRD-DDAELAAYNAMLAQLAGRDKPGEGSATG
MAB_1557|M.abscessus_ATCC_1997 VQWSRSDERLARRQDRAADRD-DDADLAAYNAMLAKMAQHNEKTRG----
TH_1033|M.thermoresistible__bu IQWSRSDQRAARRMDRAADRD-HDAELAAYNAMLAELTRREQQRTRTER-
Rv0102|M.tuberculosis_H37Rv TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
Mb0105|M.bovis_AF2122/97 TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
MMAR_0267|M.marinum_M AQWARQDRQVAARQDRHADSSYADDDLEAYNAMLRELARMRE--------
MUL_4847|M.ulcerans_Agy99 AQWARQDRQVAARKDRHADSSYADDDLEAYNAMLRELARMRE--------
MLBr_01998|M.leprae_Br4923 AQWARQDHRVAVREDRHADSTYADDDLNAYNAMLRELSRMRR--------
** * * : * * ** ** * :* *** ** .:: .
Mflv_2566|M.gilvum_PYR-GCK --
Mvan_4073|M.vanbaalenii_PYR-1 --
MSMEG_4702|M.smegmatis_MC2_155 --
MAV_1692|M.avium_104 QA
MAB_1557|M.abscessus_ATCC_1997 --
TH_1033|M.thermoresistible__bu --
Rv0102|M.tuberculosis_H37Rv --
Mb0105|M.bovis_AF2122/97 --
MMAR_0267|M.marinum_M --
MUL_4847|M.ulcerans_Agy99 --
MLBr_01998|M.leprae_Br4923 --