For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASDVRAVWPVLVGVGLLAGAVAAGIGALALADALTATGLPNPGPVTTYGLPFVRAAGEIAAVIAVGSFLF AAFLVAPQENGVLDVAGYRALRLGSAASAVWTVCAAMLVPLTISDVSGQPVLEHLSPVDVWSATSLVDIA GAWRWTAILAAAVTIASLPVLRWGWTPVLFAGSLVTLVPLALTGHSSSGGAHDLATNSLFIHLVAGAVWA GGLLALLAHAMRGRAGEGAATALAARRFSALALWCFIAMGVSGVINALVRMSPADLFRTSYGWLIVGKIV ALVVLGALGFLQRRSAVSALQADPTARGPLIRLALTEGAIFGVTFGIAVGLGRTPPPPIKRIPSAAEVAI GYDFAGPPTVARVLFDWRFDLLFGTAAIVGALVYLAALYRLRHRGDAWPVGRTVAWLLGCATLLFTTSSG LGRYMPAMFSMHMIAHMLLSMLVPILLVLGAPVTMALRALPAAGRGQPPGPREWLLAALHSKISQFFTNP VIATVMFVVGFYGLYFGGIFDAAVSNHAAHVMMNVHFLLSGYLFYWVVIGIDPTPRPIPHLGKIGMVFAS LPLHAFFGVVMMGMKTVLGEPFYRSLQLSWHTDLLGDQKLGGGVAWAAGEVPLVIVMLALLIQWRRSDDR TAKRLDRAADRDDDADLAAYNRMLAHLARRDNPPQ*
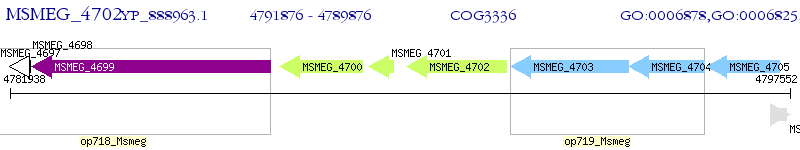
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4702 | - | - | 100% (666) | ABC-type transporter, permease components |
| M. smegmatis MC2 155 | MSMEG_6437 | - | 6e-14 | 29.85% (325) | copper resistance protein D |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0105 | - | 1e-115 | 37.07% (661) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2566 | - | 0.0 | 73.38% (665) | copper resistance D domain-containing protein |
| M. tuberculosis H37Rv | Rv0102 | - | 1e-115 | 37.07% (661) | integral membrane protein |
| M. leprae Br4923 | MLBr_01998 | - | 1e-111 | 35.78% (654) | hypothetical protein MLBr_01998 |
| M. abscessus ATCC 19977 | MAB_1557 | - | 0.0 | 64.33% (656) | copper resistance D precursor |
| M. marinum M | MMAR_0267 | - | 1e-119 | 37.86% (663) | integral membrane protein |
| M. avium 104 | MAV_1692 | - | 0.0 | 71.41% (661) | ABC-type transporter, permease components |
| M. thermoresistible (build 8) | TH_1036 | - | 1e-156 | 72.11% (380) | ABC-type transporter, permease components |
| M. ulcerans Agy99 | MUL_4847 | - | 1e-117 | 37.83% (674) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4073 | - | 0.0 | 74.39% (656) | copper resistance D domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4702|M.smegmatis_MC2_155 --------MA----SDVR--AVWPVLVGVGLLAG-AVAAGIGALALADAL
TH_1036|M.thermoresistible__bu -----MTSVG----SRVRSSAAWPVLVGVALLAG-TVAAGVAAMSLVDAL
Mflv_2566|M.gilvum_PYR-GCK -----MTSTAPHVRSGVRRAPAWAVLYRVAALAG-VTAVALAALSLADAL
Mvan_4073|M.vanbaalenii_PYR-1 -----MTSVAPVAPGGVRRAPVWAVLYGVAVLAG-VTAAALSALSLADAL
MAV_1692|M.avium_104 MTVAPPAGTASTEAVPQHRTQVWPVLAGVAVLAG-CTAAGIGTLSLASAL
MAB_1557|M.abscessus_ATCC_1997 -----MTTAP---TARKTDSPIWTIVVGLAVLAG-LVAATIGALSLAEAL
Mb0105|M.bovis_AF2122/97 MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
Rv0102|M.tuberculosis_H37Rv MGTHGATKSATSAVPTPRSNSMAMVRLAIGLLGVCAVVAAFGLVSGARRY
MMAR_0267|M.marinum_M MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
MUL_4847|M.ulcerans_Agy99 MDTDGPGVVASSAAPPSRSGSLTSVWLLLGLLAISAVLASYGVVSGVRRY
MLBr_01998|M.leprae_Br4923 MQTDEPTGTATSAIRIPPLISLRTTLMVAGLLGNVVAVAAYGLFSRTERY
. *. . . . .: .
MSMEG_4702|M.smegmatis_MC2_155 TATG---------LPNPGPVTTYGLPFVRAAGEIAAVIAVGSFLFAAFLV
TH_1036|M.thermoresistible__bu TATG---------LPNPGPVTTYGLPFVRAAGEIAAVIAVGSFMFAAFLV
Mflv_2566|M.gilvum_PYR-GCK TATG---------LPDPGPVTSYGLPFVRAAGEIAAVVAVGGFLFAAFMV
Mvan_4073|M.vanbaalenii_PYR-1 TATG---------LPDPGPVTSYGLPFVKAVGEIAAVTATGGFLFAAFMV
MAV_1692|M.avium_104 TATG---------LPDPGPVTTLGLPFLRAAGEIAAVLAVGSFLFAAFLV
MAB_1557|M.abscessus_ATCC_1997 LATG---------LPNPGPVTSYGLPFVRAAAEIAAVVAVGAFLLAAFFV
Mb0105|M.bovis_AF2122/97 AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
Rv0102|M.tuberculosis_H37Rv AEAG---------NPYPGAFVSVAEPVGFFAASLAGALCLGALIHVVMTA
MMAR_0267|M.marinum_M ATVG---------NPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
MUL_4847|M.ulcerans_Agy99 ATVGNPYPDATVGNPYPGWFLSAAEPTGYFLASLSGAVCLGGLIYVVMMS
MLBr_01998|M.leprae_Br4923 GAVG---------NPDPGSFIGVAEPVGYFGASLSSALCLGALIYFVMMV
.* * ** . . * ..::.. . *.:: .:
MSMEG_4702|M.smegmatis_MC2_155 APQENGVLDVAGYRALRLGSAASAVWTVCAAMLVPLTISDVSGQPVLEHL
TH_1036|M.thermoresistible__bu PPQPNGVLDVSGYRALQTGSVAAGVWAVCAALMVPLTVSDVSGQPVFTIL
Mflv_2566|M.gilvum_PYR-GCK PPQHNQVLDAAGYRALRMGTAASGIWTVCAVLMVPLTVSDVSGQPLSTRM
Mvan_4073|M.vanbaalenii_PYR-1 PPQQNQVLDAAGYRALRTGTAAAGAWAVCAVLMVPLTVSDVSGQPLATRL
MAV_1692|M.avium_104 PPQPSGVLDADGYRALRLGTVASGVWAVCAALLVPLTLSDVSGHPVAD-L
MAB_1557|M.abscessus_ATCC_1997 PPQTSGVLDVDGYRALRLGTAAATVLAVLSAVMVPLSVSDVSGQPLSD-F
Mb0105|M.bovis_AF2122/97 KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
Rv0102|M.tuberculosis_H37Rv KPEPDGLIDAAAFRIHLLAERVSGLWLGLAATMVVIQAAHDTGVGPARLL
MMAR_0267|M.marinum_M RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
MUL_4847|M.ulcerans_Agy99 RPEHDGLIDAGAFRIHLVTERVSVAWLVLAASMVVVQGAHDSGVSPIRLL
MLBr_01998|M.leprae_Br4923 WPQADGLIDAQAFRIHLAEERVSIGWLAFALTMVVVQAANDSGVTPAELR
*: . ::*. .:* .: : :* : :. :*
MSMEG_4702|M.smegmatis_MC2_155 SPVDVWSATSLVDIAGAWRWTAILAAAVTIASLPVLRWGWTPVLFAGSLV
TH_1036|M.thermoresistible__bu EPVSLWAAADQVNTAGAWRWTAFLAAALTLAARPVLRWGWTPFLFAGALI
Mflv_2566|M.gilvum_PYR-GCK SPADIWSVADLIDIAGAWRWTALFAALVTVVSIPVLRWSWTPVLLASSLI
Mvan_4073|M.vanbaalenii_PYR-1 SPADIWSVADLIEIAGAWRWTALLAVLVTVVSIPVLRWSWTPVLFAGSLL
MAV_1692|M.avium_104 RPAQMWSLAGLITTASAWRWTAILAAAITLASLPVLRWSWAPVLVAASLT
MAB_1557|M.abscessus_ATCC_1997 GPGELWTLASEIETVNAWRWMAFIAAGVAITSRVVLRWSWTPLLVLGSVA
Mb0105|M.bovis_AF2122/97 ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
Rv0102|M.tuberculosis_H37Rv ASGALSDSVAASEMARGWIVAAICALVVATALRLYTRWLGHVVLLVPTVL
MMAR_0267|M.marinum_M ASGAVRDAIATSEIARAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
MUL_4847|M.ulcerans_Agy99 ASGAVRDAIATSEIAHAWIVVTICALVVAVALRLNTRWVAHVVLLIPTVI
MLBr_01998|M.leprae_Br4923 TGAALFNAITTSETSRAWIVVTICALLVTLTLRVIIRYSDHFVLLIPAVI
: .* :: * :: . *: .*. ::
MSMEG_4702|M.smegmatis_MC2_155 TLVPLALTGHSSSGGAHDLATNSLFIHLVAGAVWAGGLLALLAHAMRGRA
TH_1036|M.thermoresistible__bu TLLPLGLTGHSAAGGDHDLATNSLLVHLFVAAVWAGGLLALLAHATR---
Mflv_2566|M.gilvum_PYR-GCK TLLPIALSGHSSSGGAHDIATNSLTAHMLAGALWAGGLLALLVHALRGSD
Mvan_4073|M.vanbaalenii_PYR-1 TLLPLALSGHSSSGGSHDIATNSLLIHLVAGAIWAGGLLALLVHTLRGGG
MAV_1692|M.avium_104 TLIPLGLTGHSSAGGSHDLATNGLLIHLVAAALWSGGLLALLAYALRGGQ
MAB_1557|M.abscessus_ATCC_1997 TLMPLALVGHSATGGAHDLGTNSLIIHLVSAALWAGGLVSLLIHALRGGG
Mb0105|M.bovis_AF2122/97 AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
Rv0102|M.tuberculosis_H37Rv AVVATAVTGNPGQGPDHDYATSAAIVFAVAFATLTG---LKIAAALAGTT
MMAR_0267|M.marinum_M AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
MUL_4847|M.ulcerans_Agy99 AVIATAVTGNAGQGPDHDFATSAAIVFALAAATLTG---LKAATALTGAT
MLBr_01998|M.leprae_Br4923 GVIATAVTGNAGQGPDHDYATSAVIVFALAIAALSG---LKITAALTEVT
::. .: *:.. * ** .*.. . . * :* :
MSMEG_4702|M.smegmatis_MC2_155 GEGAATALAARRFSALALWCFIAMGVSGVINALVRMSPADLFRTSYGWLI
TH_1036|M.thermoresistible__bu -TADHADLAARRFSAVAFWCYIAMALSGVINALVRMNVSDLLSTNYGRLV
Mflv_2566|M.gilvum_PYR-GCK DGDDHAALAARRFSAVALWCFVAMALSGTINALVRVRPADLVTTTYGTLL
Mvan_4073|M.vanbaalenii_PYR-1 ----HGPLAARRFSAVALWCFVAMAFSGVINALVRIRPADLVSTQYGLLI
MAV_1692|M.avium_104 -GGGHLGLATRRFSAIALWCWVAMALSGLVNAAVRVQPSDLLATGYGRLV
MAB_1557|M.abscessus_ATCC_1997 ----HLDVAARRFSMLALWCYVAIGLSGIINALIRVQLSDLFTTRYGWLL
Mb0105|M.bovis_AF2122/97 --------PSRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFARLG
Rv0102|M.tuberculosis_H37Rv --------PSRAVLVTQVTCGALALAYGAMLLYLFIPG-WAVDSDFARLG
MMAR_0267|M.marinum_M --------PTRTVQLMQVACGALAVSYGAVLLYLFVPG-WDFGSEFARIG
MUL_4847|M.ulcerans_Agy99 --------PTRAVQLMQVACGALAVSYGTVLLYLFVPG-WDFGSEFARIG
MLBr_01998|M.leprae_Br4923 --------PSRAVLIVQVVCGVLALAYGAELVYLLSPGAAMIDSDFGRLG
.:* . . * * : . : :. :
MSMEG_4702|M.smegmatis_MC2_155 VGKIVALVVLGALGFLQRRSAVSALQAD---PTARGPLIRLALTEGAIFG
TH_1036|M.thermoresistible__bu LAKIVAFALLGVIGWQQRRRGVAAVQAD---PDSRGPLIRLALTEAVIFG
Mflv_2566|M.gilvum_PYR-GCK LAKIAALVALGVLGWQQRRRGVAALQAD---PRARGPLIRLALVEAAVFG
Mvan_4073|M.vanbaalenii_PYR-1 LAKLVALVTLGGLGWQQRRRAVAALQQD---PDARQPLIRLALVEAVVFG
MAV_1692|M.avium_104 LAKAAALCLLGGVGWRQRRVNVAALQAVSTLARARRALLRLTLIEAALFG
MAB_1557|M.abscessus_ATCC_1997 MGKATALLLLGVLGYLQRRSAITALDED---PENRQPLIRLAGVEAIIFA
Mb0105|M.bovis_AF2122/97 LLAGVILTSVWLFDCWRLLVRPPHAGRRR----------GGGSGAALAMM
Rv0102|M.tuberculosis_H37Rv LLAGVILTSVWLFDCWRLLVRPPHAGRRR----------GGGSGAALAMM
MMAR_0267|M.marinum_M LLAGLVLVLVWVFDCWQLFVRPPRSTR-------------GATAAALAMM
MUL_4847|M.ulcerans_Agy99 LLAGLLLVLVWVFDCWQLFVRPPRSTR-------------GATAAALAMM
MLBr_01998|M.leprae_Br4923 LVAGVLVTLVCFSDCWALVMGRPHSDHR-------------TRLTALAMM
: . : . . . :
MSMEG_4702|M.smegmatis_MC2_155 VTFGIAVGLGRTPPPPIKRI-PSAAEVAIGYDFAGPPTVARVLFDWRFDL
TH_1036|M.thermoresistible__bu ATFGIAVGLGRTPPPPPPRQ-PSPVEVAIGYDLAGPPTVARILFDWRFDL
Mflv_2566|M.gilvum_PYR-GCK LTFGVAVALGRTPPPPPLNPNLSIPAVEIGYDLDGPPTLGRLLFDWRFDL
Mvan_4073|M.vanbaalenii_PYR-1 LTFGVAVALGRTPPPPPTNTNLSIPAVEIGYDLAGPPTVGKVLFDWRFDL
MAV_1692|M.avium_104 LTFGIAVGLGRTAPPPPPARLPSIAEAEIGYDFDGPPTLTRILFDWRFDL
MAB_1557|M.abscessus_ATCC_1997 VTFGIAVGLGRTPPPPPVNLNPSPVEVAIGYNLDGPPTPGRLMFDWRFDL
Mb0105|M.bovis_AF2122/97 AAMASIAAMAVMTAPRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
Rv0102|M.tuberculosis_H37Rv AAMASIAAMAVMTAPRFLTHAFTAWDVFLGYELPQPPTIARVLTVWRFDS
MMAR_0267|M.marinum_M AVLAAIGAMAVQTAPRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
MUL_4847|M.ulcerans_Agy99 AVLAAIGAMAVQTAPRFVAHQFTAWDVFLGYELPQPPDVGRLITLWRFDS
MLBr_01998|M.leprae_Br4923 AVTAATAAMSVQTAPRFLTHRFTAWDVFLGYELPQPPNIVRFFTVWRFDS
. . .:. ..* : . :**:: ** :.: ****
MSMEG_4702|M.smegmatis_MC2_155 LFGTAAIVGALVYLAALYRLRHRGDAWPVGRTVAWLLGCATLLFTTSSGL
TH_1036|M.thermoresistible__bu IFGPAAXX------------------------------------------
Mflv_2566|M.gilvum_PYR-GCK VLGTAAIVLALVYLAGVRRLRRRGDAWPTGRVVAWLLGCLVLLLATSSGF
Mvan_4073|M.vanbaalenii_PYR-1 IFGTAAIVFALVYLAGVRRLRRRGDAWPIGRVVAWLLGCLVLLLATSSGI
MAV_1692|M.avium_104 IFGTAAIVLAGLYVAGVVRLRRRGDRWQPGCTSSWLLGCLVLLFVTSSGV
MAB_1557|M.abscessus_ATCC_1997 IFGTAAIVFAIAYVVGVRRLKQRGDEWPIGRTISWLFGCGFLLIATSSGV
Mb0105|M.bovis_AF2122/97 LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
Rv0102|M.tuberculosis_H37Rv LIGAAGVVLAIGYAAGFAALRRRGNSWPVGRLIAWLTGCAALVFTSGSGV
MMAR_0267|M.marinum_M LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
MUL_4847|M.ulcerans_Agy99 LVGVAGVVLAAGYVAGFVRLRRRGHAWPVSRLVSWLIGCAVLVITSSSGV
MLBr_01998|M.leprae_Br4923 LIGATAIVLGIGYVAGFVRVRRAGNTWPVGRLVAWLTGCVALVFTSSSGV
:.* :.
MSMEG_4702|M.smegmatis_MC2_155 GRYMPAMFSMHMIAHMLLSMLVPILLVLGAPVTMALRALPAAGRGQPPGP
TH_1036|M.thermoresistible__bu -XLLPGQFGA----------------------------------------
Mflv_2566|M.gilvum_PYR-GCK GRYMPAMFSIHMIAHMMLSMLAPILLVLGAPVTLALRALPVAGRTGAPGP
Mvan_4073|M.vanbaalenii_PYR-1 GRYMPAMFSMHMIAHMLLSMLAPILLVLGAPVTLALRALPAAGRDGPPGP
MAV_1692|M.avium_104 GRYMPAMFSMHMVVHMCLSMLVPILLALGAPVTLALRALPAAGRGDPPGP
MAB_1557|M.abscessus_ATCC_1997 GRYMPAMFSMHMVAHMMLSMLVPVLLVLGGPVTLLLRVLPAAGKSDPPGL
Mb0105|M.bovis_AF2122/97 RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
Rv0102|M.tuberculosis_H37Rv RAYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRVLPVTGDGRPPGA
MMAR_0267|M.marinum_M RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAGEGQPPGP
MUL_4847|M.ulcerans_Agy99 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALPAAVEGQPPGP
MLBr_01998|M.leprae_Br4923 RTYGSAMFSVHMAEHMTLNMFIPVLLVLGGPVTLALRALSAAGDGQQPGP
.. *.
MSMEG_4702|M.smegmatis_MC2_155 REWLLAALHSKISQFFTNPVIATVMFVVGFYGLYFGGIFDAAVSNHAAHV
TH_1036|M.thermoresistible__bu --------------------------------------------------
Mflv_2566|M.gilvum_PYR-GCK REWLLAALHSRVSQFLTNPFVATGLFVVGFYGLYFSGLFDAAAGSHAAHV
Mvan_4073|M.vanbaalenii_PYR-1 REWLLAALHSRYSQVVTNPIVATALFVVGFYGLYFSGLFEAAAGSHFAHL
MAV_1692|M.avium_104 REWLLAALHSRFSRLLTNPVVATVLFVAGFYGLYLSNLFDTTASSHAGHL
MAB_1557|M.abscessus_ATCC_1997 REWVQLALHSKFSRFLTHPLVATSLFIGGFYGLYLSGLYDAAVDVHAAHL
Mb0105|M.bovis_AF2122/97 REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
Rv0102|M.tuberculosis_H37Rv REWLTWLLHSRVTTFLSHPITAFVLFVASPYIVYFTPLFDTFVRYHWGHE
MMAR_0267|M.marinum_M REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
MUL_4847|M.ulcerans_Agy99 REWLTWLLHSRVTAFFSHPIIAFILFVASPYIVYFTPLFDTLVRYHWGHE
MLBr_01998|M.leprae_Br4923 REWLTWLMHSSVTAFLSHPVTNFILFVGSPYIVYFTPLFDTLVRYHWGHE
MSMEG_4702|M.smegmatis_MC2_155 MMNVHFLLSGYLFYWVVIGIDPTPRPIPHLGKIGMVFASLPLHAFFGVVM
TH_1036|M.thermoresistible__bu --------------------------------------------------
Mflv_2566|M.gilvum_PYR-GCK AMNVHFLLSGYLFYWVVIGVDPTPRPIPHLAKLGMVFASLPLHAFFGVVL
Mvan_4073|M.vanbaalenii_PYR-1 AMNLHFLVSGYLFYWVVIGIDPTPRPIPHLAKLAVVFASLPLHAFFGVVL
MAV_1692|M.avium_104 LMNLHFLLSGYLFYWVVIGVDPTPRPIPPLAKLAVVFASLPLHAFFGVVL
MAB_1557|M.abscessus_ATCC_1997 AMNLHFLLSGYLFYWVVIGIDPSPRRLPPVAKLGIVLVSLPLHAFFGVIL
Mb0105|M.bovis_AF2122/97 FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
Rv0102|M.tuberculosis_H37Rv FMAIHFLVVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MMAR_0267|M.marinum_M FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MUL_4847|M.ulcerans_Agy99 FMAVHFLIVGYLFYWAIIGIDPGPRRLPYPGRIGLLFAVMPFHAFFGIAL
MLBr_01998|M.leprae_Br4923 FMAIHFLLVGYIFYWAIIGIDPGPRRLPYLARIGLLFAVMPFHAFFGIAL
MSMEG_4702|M.smegmatis_MC2_155 MGMKTVLGEPFYRSLQLSWHTDLLGDQKLGGGVAWAAGEVPLVIVMLALL
TH_1036|M.thermoresistible__bu --------------------------------------------------
Mflv_2566|M.gilvum_PYR-GCK MGTQSVLAESFYRSLQLPWHTDLLADQRVGGGIAWAAGEVPLVVVLIALF
Mvan_4073|M.vanbaalenii_PYR-1 MGTQSVLAESFYRSLQLSWHTDLMGDQKLGGGIAWAAGEIPLVVVLIALF
MAV_1692|M.avium_104 MGTRKVLGADYYRSLGFSWHTDLLGDQRLGGGIAWSAGEFPLVIVMLALL
MAB_1557|M.abscessus_ATCC_1997 MGTKSILGEKFYSNLALPWRIDLAADQHMGGAITWATGELPLMVVMIALV
Mb0105|M.bovis_AF2122/97 MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
Rv0102|M.tuberculosis_H37Rv MTMSSTVGATFYRSVNLPWLSSIIADQHLGGGIAWSLTELPVIMVIVALV
MMAR_0267|M.marinum_M MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
MUL_4847|M.ulcerans_Agy99 MTMNSAIGDTFYRSVNLPWLASIESDQHLGGGIAWGATELPIIIVIVALV
MLBr_01998|M.leprae_Br4923 MTMTSAISAEFYRSVNLPWLASIDADQHIGGAIAWSATELPVIIVIVALM
MSMEG_4702|M.smegmatis_MC2_155 IQWRRSDDRTAKRLDRAAD-RDDDADLAAYNRMLAHLARRDNPPQ-----
TH_1036|M.thermoresistible__bu --------------------------------------------------
Mflv_2566|M.gilvum_PYR-GCK IQWRRTDDRTARRLDRAAD-RDDDADLAAYNNMLAELARRERERG-----
Mvan_4073|M.vanbaalenii_PYR-1 VQWRRTDDRTAKRLDRAAD-RDDDADLAAYNAMLAELARRERERS-----
MAV_1692|M.avium_104 VQWARSDRRTAKRLDRAAD-RDDDAELAAYNAMLAQLAGRDKPGEGSATG
MAB_1557|M.abscessus_ATCC_1997 VQWSRSDERLARRQDRAAD-RDDDADLAAYNAMLAKMAQHNEKTRG----
Mb0105|M.bovis_AF2122/97 TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
Rv0102|M.tuberculosis_H37Rv TQWARQDRRVASREDRHADSDYADDELEAYNAMLRELSRMRR--------
MMAR_0267|M.marinum_M AQWARQDRQVAARQDRHADSSYADDDLEAYNAMLRELARMRE--------
MUL_4847|M.ulcerans_Agy99 AQWARQDRQVAARKDRHADSSYADDDLEAYNAMLRELARMRE--------
MLBr_01998|M.leprae_Br4923 AQWARQDHRVAVREDRHADSTYADDDLNAYNAMLRELSRMRR--------
MSMEG_4702|M.smegmatis_MC2_155 --
TH_1036|M.thermoresistible__bu --
Mflv_2566|M.gilvum_PYR-GCK --
Mvan_4073|M.vanbaalenii_PYR-1 --
MAV_1692|M.avium_104 QA
MAB_1557|M.abscessus_ATCC_1997 --
Mb0105|M.bovis_AF2122/97 --
Rv0102|M.tuberculosis_H37Rv --
MMAR_0267|M.marinum_M --
MUL_4847|M.ulcerans_Agy99 --
MLBr_01998|M.leprae_Br4923 --