For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
WTRQATVASVNAPDALTDDQRQPLRIEIVVVLMVTFALSAYTAILDLIEAVLLGLAGQTVALNPRRSSFD LIDLGLNLATVFQLVAWGFLAVYLLWRSGFGPAAIGLGRPRYRLDLLGGIGLAALIGLPGLALYVTARAL GLSASVVPSELNDTWWRIPVLILVSFANAWAEEIIVVGFLLTRLHQLRVRPATALVLSSLLRGTYHLYQG FSAGLGNVVMGLVFGYAWRRTGRLWPLIIAHGVINTVAYVGYALVADRLNWLY*
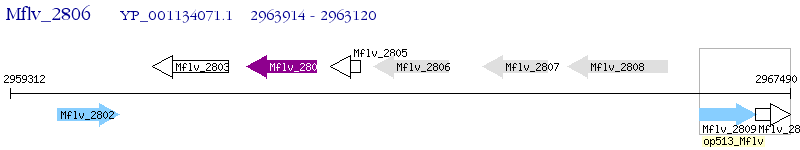
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2806 | - | - | 100% (264) | abortive infection protein |
| M. gilvum PYR-GCK | Mflv_1143 | - | e-104 | 73.09% (249) | abortive infection protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0129c | - | 1e-93 | 65.00% (240) | abortive infection protein |
| M. marinum M | MMAR_0147 | - | 2e-07 | 28.50% (207) | hypothetical protein MMAR_0147 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6420 | - | 1e-102 | 76.03% (242) | abortive infection protein |
| M. thermoresistible (build 8) | TH_1470 | - | 1e-104 | 72.69% (249) | abortive infection protein |
| M. ulcerans Agy99 | MUL_4950 | - | 5e-06 | 28.88% (187) | hypothetical protein MUL_4950 |
| M. vanbaalenii PYR-1 | Mvan_3716 | - | 1e-141 | 96.89% (257) | abortive infection protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2806|M.gilvum_PYR-GCK MWTRQATVASVNAPDALTDDQRQPLRIEIVVVLMVTFALSAYTAILDLIE
Mvan_3716|M.vanbaalenii_PYR-1 -------MASVNAPDALTDDQRRPLQIEIVVVLMVTFALSAYTAILDLID
MSMEG_6420|M.smegmatis_MC2_155 ----MTGPLEHREHSEHRQPSRRTIKIEIVVVLAVTFGLSAYTALLRLIE
TH_1470|M.thermoresistible__bu ----VSGPAE-----ELTAPQRRALRLEIAVVLLVTFGLTAYSALLRLTE
MAB_0129c|M.abscessus_ATCC_199 --------------MAESEMSARTIRLEIAIVLAISFGMSAFSAILQFAS
MMAR_0147|M.marinum_M -----------MLPTSSSTPDAPALRPTAAPAHRWGLGAFVLVELVYLLS
MUL_4950|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2806|M.gilvum_PYR-GCK AVLLGLAGQTVALNPRRSSFDLIDLGLNLATVFQLVAWGFLAVYLLWRSG
Mvan_3716|M.vanbaalenii_PYR-1 AVLLGLAGQTVALNPRRSSFDLIDLGLNLATVFQLVAWGFLAVYLLWRSG
MSMEG_6420|M.smegmatis_MC2_155 AVLLGLSGQTVALNPRRSPFDLIDLGLHLAGVFNLLAWGALALYLLWRSG
TH_1470|M.thermoresistible__bu SVLLGLSGQIVALNPRRSQFSLIDLGLNLAAVFQLMGWGALALYLLWRSG
MAB_0129c|M.abscessus_ATCC_199 AVLMGLSGQRVALNPRRAELSLIDLGLNLVSITRLVAWGALAVYLLWRSG
MMAR_0147|M.marinum_M STLLALV--VASAGPR--SAALISLAVAAPTVIAAG----LAVFITMRRG
MUL_4950|M.ulcerans_Agy99 --MLALV--VASAGPR--SAALISLAVAAPTVIAAG----LAVFITMRRG
::.* .: .** **.*.: : **::: * *
Mflv_2806|M.gilvum_PYR-GCK FGP-AAIGLG--RPRYRLDLLGGIG-LAALIGLPGLALYVTARALGLSAS
Mvan_3716|M.vanbaalenii_PYR-1 FGP-GAIGLG--RPRYRLDLLGGIG-LAALIGLPGLALYVTARALGLSAS
MSMEG_6420|M.smegmatis_MC2_155 IGP-KAIGLG--RPQARADGLGGLA-LAALIGLPGLGFYVVARILGLSAD
TH_1470|M.thermoresistible__bu SGP-ARIGLG--RLRWRPDLLGGLG-LAALIGLPGIGLYLAARALGLSAA
MAB_0129c|M.abscessus_ATCC_199 FRP-SSVGLG--RWRWRADGLGALG-LAALIGLPGLALYVGARWMGMSVQ
MMAR_0147|M.marinum_M NGPRTDLRLGGTWRDVRLGLVFGLGGLVVSVPASMLYASITGPDANSALY
MUL_4950|M.ulcerans_Agy99 NGPRTDLRMGGTWRDVRLGLVFGLGGLVVSVAASMLYASITGPDANSALY
* : :* * . : .:. *.. : . : : . . :
Mflv_2806|M.gilvum_PYR-GCK VVPSELNDTW-WRIPVLILVSFANAWAEEIIVVGFLLTRLHQLRVRPATA
Mvan_3716|M.vanbaalenii_PYR-1 VVPSELNDTW-WRIPILILVSFANAWAEEIIVVGFLLTRLHQLRVRPVTA
MSMEG_6420|M.smegmatis_MC2_155 VEPAELYDTW-WRIPVLLGSAFANGWAEEIIVVGFLLTRLRQLDISPTRA
TH_1470|M.thermoresistible__bu VEPAELYDTW-WRIPVLLAIAFANGWAEEIVVVGFLVTRLRQLRVTPVAA
MAB_0129c|M.abscessus_ATCC_199 VIPASLDDTW-WRLPVLVLSAFANGWAEEVVVIAYLQTRLHQLGYGTAAA
MMAR_0147|M.marinum_M KVFGDVRASWPWAVAVFIVVVFVGPLCEEILYRGLLWGALERRWGQWVAL
MUL_4950|M.ulcerans_Agy99 KVFGDVRASWPWAVAVFIVVVFVGPLCEEILYRGLLWGALERRWGQWVAL
..: :* * :.::: *.. .**:: . * *.: .
Mflv_2806|M.gilvum_PYR-GCK LVLSSLLRGTYHLYQGFSAGLGNVVMGLVFGYAWRRTGRLWPLIIAHGVI
Mvan_3716|M.vanbaalenii_PYR-1 LVLSSLLRGTYHLYQGFSAGLGNVVMGLVFGYTWQRTGRLWPLIIAHGVI
MSMEG_6420|M.smegmatis_MC2_155 LVLSALLRGAYHLYQGYSAGPGNLVMGLVFGYAWQRTGRLWPLIIAHTLI
TH_1470|M.thermoresistible__bu VLASSLLRGAYHLYQGFSAGLGNLVMGLIFGWLYLRTGRLWPLIVAHAVI
MAB_0129c|M.abscessus_ATCC_199 IVSSALLRGCYHLYQGVSAGVGNLAMGVVFGYVWHRTGRLWPLVVAHGVI
MMAR_0147|M.marinum_M VVSTAVFALAHFEFT---RAPLLLVIAVPIALARLYSGGLWASIVAHQVT
MUL_4950|M.ulcerans_Agy99 VVSTAVFALAHFEFT---RAPLLLVIAVPIALARLYSGGLWASIAAHQVT
:: :::: :. : . :.:.: :. :* **. : ** :
Mflv_2806|M.gilvum_PYR-GCK NTVAYVGYALVADRLNWLY
Mvan_3716|M.vanbaalenii_PYR-1 NTVAYVGYALVADRLNWLY
MSMEG_6420|M.smegmatis_MC2_155 DAVAFVGYALLAQHLGWLR
TH_1470|M.thermoresistible__bu DAVAFVGYALAADHLTWLT
MAB_0129c|M.abscessus_ATCC_199 DTVAFVGYALLRDHLGWLH
MMAR_0147|M.marinum_M NSLPGLVLMLILTGTMPAS
MUL_4950|M.ulcerans_Agy99 NSLPGLVLMLILTGTMPAS
:::. : *