For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SASLGAAPAGPPSPPPRLDHRPSRDPALAGLRAVAALLVVGTHAAFATGYLTHGYLGTMYARLEIGVAVF FVLSGFLLFRPWVAAAAEGRRGPSVRRFARRRLRRIVPAYLITVVAVFEVYTVFTPGPNPGQTWTGLLGH LTFTHIYA*
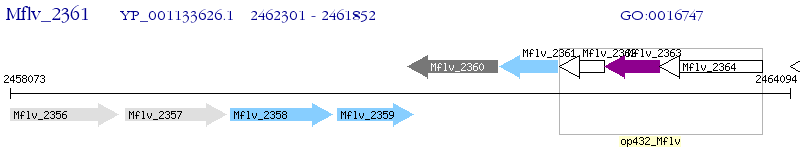
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2361 | - | - | 100% (149) | acyltransferases-like protein |
| M. gilvum PYR-GCK | Mflv_2226 | - | 1e-38 | 61.29% (124) | acyltransferase 3 |
| M. gilvum PYR-GCK | Mflv_0420 | - | 3e-18 | 44.26% (122) | acyltransferase 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1286 | - | 5e-40 | 62.10% (124) | acyltransferase |
| M. tuberculosis H37Rv | Rv1254 | - | 5e-40 | 62.10% (124) | acyltransferase |
| M. leprae Br4923 | MLBr_01101 | - | 1e-38 | 59.68% (124) | putative acyltransferase |
| M. abscessus ATCC 19977 | MAB_1404 | - | 2e-32 | 54.96% (131) | acyltransferase |
| M. marinum M | MMAR_4188 | - | 1e-37 | 57.26% (124) | integral membrane acyltransferase |
| M. avium 104 | MAV_1402 | - | 8e-37 | 56.69% (127) | putative acyltransferase, putative |
| M. smegmatis MC2 155 | MSMEG_6230 | - | 8e-44 | 64.57% (127) | acyltransferase |
| M. thermoresistible (build 8) | TH_1720 | - | 1e-43 | 59.03% (144) | PROBABLE ACYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_4492 | - | 2e-37 | 57.26% (124) | integral membrane acyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4285 | - | 5e-65 | 80.26% (152) | acyltransferase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01101|M.leprae_Br4923 -----MTLSEERDAQSGLKQVSHVDRVASLTGIRAVAALLIVGTHAAYTT
MAV_1402|M.avium_104 -----MTLSEEQDAQGGLEQTSRVDRVASLTGVRAVAALLVVGTHAAYTT
MMAR_4188|M.marinum_M -----MTLPEATDAQGGLEQISHVDRVASLTGIRAVAALLVVATHAAYTT
MUL_4492|M.ulcerans_Agy99 -----MTLPEATDAQGGLEQISHVDRVASLTGIRAVAALLVVTTHAAYTT
Mb1286|M.bovis_AF2122/97 -----MTLPKERAAQGGLERIAHVDRVASLTGIRAVAALLVVGTHAAYTT
Rv1254|M.tuberculosis_H37Rv -----MTLPKERAAQGGLERIAHVDRVASLTGIRAVAALLVVGTHAAYTT
MAB_1404|M.abscessus_ATCC_1997 -----MTAVSPTESAP---VVRASGRVAALTGVRAVAATLVVATHAAYST
MSMEG_6230|M.smegmatis_MC2_155 MAAAPGPDPTDGADQGGLEQVSRADRIASLTGVRAVAALTVMGTHAAYGT
TH_1720|M.thermoresistible__bu ----VRPSPETDLDQGGLEQVSTVDRVAALTGVRAVAALSVMGTHAAYGT
Mflv_2361|M.gilvum_PYR-GCK -MSASLGAAPAGPPSPPPRLDHRPSRDPALAGLRAVAALLVVGTHAAFAT
Mvan_4285|M.vanbaalenii_PYR-1 -MSASLGVRCGAAPAGPP--DHTSPRDPALTGLRAVAALLVVSTHAAFAT
* .:*:*:***** :: ****: *
MLBr_01101|M.leprae_Br4923 GKYTHG-------YWGLVGARLEIGVPIFFALSGFLLFSPWVKSAATGSP
MAV_1402|M.avium_104 GKYTHG-------YWGLVGSRMEIGVPIFFVLSGYLLFRPWVKSAATGGP
MMAR_4188|M.marinum_M GKYTHG-------YWGLVSARMEIGVPIFFVLSGFLLFRPWVHSAVTGAP
MUL_4492|M.ulcerans_Agy99 GKYTHG-------YWGLVSARMEIGVPIFFVLSGFLLFRPWVHSAVTGAP
Mb1286|M.bovis_AF2122/97 GKYTHG-------YWGLMSSRMEIGVPIFFVLSGFLLFRPWVKSAATGGP
Rv1254|M.tuberculosis_H37Rv GKYTHG-------YWGLMSSRMEIGVPIFFVLSGFLLFRPWVKSAATGGP
MAB_1404|M.abscessus_ATCC_1997 GNYVVDGVSPWKAYLHLLQSRLEIGVPIFFVLSGFLLFLPWVRAAADNTA
MSMEG_6230|M.smegmatis_MC2_155 GLLTHG-------YVGLLGARLEIGVAIFFVLSGLLLFRPWVRAVATGAQ
TH_1720|M.thermoresistible__bu GTYNHG-------YVGLIFARMDIGVAVFFVLSGFLLFRPWVRAAADGTA
Mflv_2361|M.gilvum_PYR-GCK GYLTHG-------YLGTMYARLEIGVAVFFVLSGFLLFRPWVAAAAEGRR
Mvan_4285|M.vanbaalenii_PYR-1 GYLLHG-------YLGTMYARLEIGVAVFFVLSGFLLFRPWVRAAAAGRR
* . * : :*::***.:**.*** *** *** :.. .
MLBr_01101|M.leprae_Br4923 PPSVSRYAWHRVRRIMPAYVITVLFAYALYHFRAAGPNPGHSWMGLVRNL
MAV_1402|M.avium_104 PPSLSRYVWHRVRRIMPPYVITVLFAYVLYHFREAGPNPGHSWLGLARNL
MMAR_4188|M.marinum_M PPSLRRYTAHRVRRIMPAYVVVVLIAYVIYHYRIAGPNPGHSWLGLVRNL
MUL_4492|M.ulcerans_Agy99 PPSLRRYTAHRVRRIMPAYVVVVLIAYVIYHYRIAGPNPGHSWMGLVRNL
Mb1286|M.bovis_AF2122/97 PPSLSRYAWHRVRRIMPAYTVTVLLAYLVYHFRTAGPNPGHTWVGLFRNL
Rv1254|M.tuberculosis_H37Rv PPSLSRYAWHRVRRIMPAYTVTVLLAYLVYHFRTAGPNPGHTWVGLFRNL
MAB_1404|M.abscessus_ATCC_1997 SPSVKRYAWHRFRRIMPAYWILVAIAYIWYHIRDQYPNPGHTWLGLFRNL
MSMEG_6230|M.smegmatis_MC2_155 APSVRRYARSRVRRIMPAYVVTVLLVFGIYELRSVEPNPGHTWTGLWRHL
TH_1720|M.thermoresistible__bu APSVRRYARNRVRRIMPAYVVTVLAAYGIYAVRELEPNPGHTWVGLLRNL
Mflv_2361|M.gilvum_PYR-GCK GPSVRRFARRRLRRIVPAYLITVVAVFEVYTVFTPGPNPGQTWTGLLGHL
Mvan_4285|M.vanbaalenii_PYR-1 APSARRYGRQRMRRIVPAYVVTVVAVFEIYTVFTPGPNPGQTWQGLLRYL
** *: *.***:*.* : * .: * ****::* ** *
MLBr_01101|M.leprae_Br4923 TLTQIYTDGYLGSYLHQGLTQMWSLAVEVAFYVALPLLAYLLLVLLCQRR
MAV_1402|M.avium_104 TLTQIYCNGYLGKYLHQGLTQMWSLAVEASFYVILPLLAYLLLVLVCRRQ
MMAR_4188|M.marinum_M TLTQIYTDGYLGSYLHQGLTQMWSLAVEASFYVLLPFLAYLLLVVISRRR
MUL_4492|M.ulcerans_Agy99 TLTQIYTDGYLGSYLHQGLTQMWSLAVEASFYVLLPFLAYLLLVVISRRR
Mb1286|M.bovis_AF2122/97 TLTQIYTDGYLGAFLHQGLTQMWSLAVEVAFYLALPALAYLLLVLVCRRR
Rv1254|M.tuberculosis_H37Rv TLTQIYTDGYLGAFLHQGLTQMWSLAVEVAFYLALPALAYLLLVLVCRRR
MAB_1404|M.abscessus_ATCC_1997 TLTQIYTSNYGYRYIHQGMTQIWSLAVEAAFYVALPFLAWLLVKKLCGGQ
MSMEG_6230|M.smegmatis_MC2_155 TLTQIYRGDYYTSYLHQGLTQMWSLAVEVTFYAALPLLAYVLLVVLCRRH
TH_1720|M.thermoresistible__bu TLTQIYSDDYLTAYAHQGLTQMWSLAVEVAFYAALPVLAWLLLVVLARRR
Mflv_2361|M.gilvum_PYR-GCK TFTHIYA-------------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 TFTHIYTDDYLVTLLHPGLSQMWSLAVEASFYVALPALAFLVLQVLCRGR
*:*:**
MLBr_01101|M.leprae_Br4923 WQPRLVLVVLAGMLLISPGWLILVHPDHWFPDGARLWLPTYLAWFLGGMI
MAV_1402|M.avium_104 WQPKLVLGALAGLALVSPAWLVLVHTDHWFPDGARLWLPTYLAWFLAGMM
MMAR_4188|M.marinum_M WQPKLVVGALLAMATITPGWLVLIHTVSLFPDGARLWLPSYLAWFLGGML
MUL_4492|M.ulcerans_Agy99 WQPKLVVGALLAMATITPGWLVLIHTVSLFPDGARLWLPSYLAWFLGGML
Mb1286|M.bovis_AF2122/97 WQPRLLLATMAGLTMISPAWLILVHNTHWMPDGARLWLPTYLAWFVGGMM
Rv1254|M.tuberculosis_H37Rv WQPRLLLATMAGLTMISPAWLILVHNTHWMPDGARLWLPTYLAWFVGGMM
MAB_1404|M.abscessus_ATCC_1997 WRPARLLVGLGGLALVTPLWMSLVYTIGAFPDGAKLWLPAYLSWFVGGMI
MSMEG_6230|M.smegmatis_MC2_155 FRPWRLLGGLAVLGAVSPLWLVASH-AGRLPVSAGAWLPHYLVWFIGGMM
TH_1720|M.thermoresistible__bu WRPVPLLTGLGVLAVVSPLWVVVLHRTDWLPLGAGFWLPHYLIWFVGGMV
Mflv_2361|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 WRPGRALVGIALLAAVTPVWLVVVTATDLLPNSAGMWLPAHLAWFAGGMA
MLBr_01101|M.leprae_Br4923 LAVLQVMGVRCYAFGAIPLAIICYFIVSTPIAGAPTTSPASLGEALIKVG
MAV_1402|M.avium_104 LTVLQEMGARAYGFVAIPLAVVSYFIVSTPIAGAPTTSPATLSEALFKAC
MMAR_4188|M.marinum_M LTVLQAMKVQCYALAALPLATICYFIVATPIAGAPTTSPSALTEAVVKAC
MUL_4492|M.ulcerans_Agy99 LTVLQAMKVQCYALAALPLATICYFIVATPIAGAPTTSPSALTEAVVKAC
Mb1286|M.bovis_AF2122/97 LAVLAAMGVRCYAFVAIPLAVICYFIVSTPIAGAPTTSPTALAEALVKTA
Rv1254|M.tuberculosis_H37Rv LAVLAAMGVRCYAFVAIPLAVICYFIVSTPIAGAPTTSPTALAEALVKTA
MAB_1404|M.abscessus_ATCC_1997 LSVLAVMGVRCYALVTVPLAVICYLIVSTRIAGDPTTSPWALPQALTKSI
MSMEG_6230|M.smegmatis_MC2_155 LAVLATMGVRCHAAAVLPVAVAGYLVVSTPIAGTTTAFTLTLGEDITKTM
TH_1720|M.thermoresistible__bu LAVLQAQGVRAYAAATVPVAVAAYLIVATPIAGDTGAPVLNLGQDLAKVM
Mflv_2361|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 LAVLEAVGARCAAIVAIPVAVGLYLAVSTSLG-GTLAGPDPAWTPIVKAL
MLBr_01101|M.leprae_Br4923 FYAAIATLAVAPLTLGDLGWYSWLLSSRPMVWLGEISYEIFLIHLVTMEF
MAV_1402|M.avium_104 FYAVIAALAVAPLALGDHGWYSRLLASRPMVWLGEISYEIFLIHLITMEF
MMAR_4188|M.marinum_M FYTVIATLAVAPLALGDQGWYSQLLASRPMVWLGEISYEIFLVHLVTMEF
MUL_4492|M.ulcerans_Agy99 FYTVIATLAVAPLALGDQGWYSQLLASRPMVWLGEISYEIFLVHLVTMEF
Mb1286|M.bovis_AF2122/97 FYAVIAVLAVAPLALGDQGWYAQLLASRPMVFLGEISYEIFLIHLVTMEI
Rv1254|M.tuberculosis_H37Rv FYAVIAVLAVAPLALGDQGWYAQLLASRPMVFLGEISYEIFLIHLVTMEI
MAB_1404|M.abscessus_ATCC_1997 FYVVIATLLVAPAALGDNGWYVRFLSTRPMVWLGEISYELFLIHMILMEI
MSMEG_6230|M.smegmatis_MC2_155 FYAVIATLMVAPLALGSTGFYARVMASRPMVWLGEISYEIFLLHVVVMDL
TH_1720|M.thermoresistible__bu FYAVIATLVVAPPALGNRGWYTRMLGSRPMVWLGEISYEIFLVHVIVMEI
Mflv_2361|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 LYAVIATLAVAPVALGSRDSYSRVLASRPMVWLGEISYEIFLVHVAVMAL
MLBr_01101|M.leprae_Br4923 AMVYVVQDHVYTGSMLNLIVATLVLTIPLAWLLHR--------CTRMRS-
MAV_1402|M.avium_104 AMDYIVGARVYTGSMLNLFVATLVLTIPLAWLLHR--------FTRVRD-
MMAR_4188|M.marinum_M AMVYVVRYRVYTGSMLNLFIATMVITIPLAWVLHR--------FTRVRSE
MUL_4492|M.ulcerans_Agy99 AMVYVVRYWVYTGSMLNLFIATMVITIPLAWVLHR--------FTRVRSE
Mb1286|M.bovis_AF2122/97 AMVDVLGYRVYTSSMVNLCLVTLVLTIPLAWLLHR--------FTRVQGD
Rv1254|M.tuberculosis_H37Rv AMVDVLGYRVYTSSMVNLCLVTLVLTIPLAWLLHR--------FTRVQGD
MAB_1404|M.abscessus_ATCC_1997 VMVDLLHYHVYTGSVVIVFLVTMLVSVPASWLIHRTVEVILARSSRLRKS
MSMEG_6230|M.smegmatis_MC2_155 VMEFVLGWHVYTGWAAGLFAATLAATIPVAWVLHR--------LTRP---
TH_1720|M.thermoresistible__bu AMVSVLRWPVYTGSVLGLFATTLALTIPVAWLLHR--------FTRPPIR
Mflv_2361|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 TMHLVLGWPLFTGSMPGLIAATLALTVPVAWLLNR--------VTRPQIK
MLBr_01101|M.leprae_Br4923 ----------------------------------------
MAV_1402|M.avium_104 ----------------------------------------
MMAR_4188|M.marinum_M ----------------------------------------
MUL_4492|M.ulcerans_Agy99 ----------------------------------------
Mb1286|M.bovis_AF2122/97 RPS-------------------------------------
Rv1254|M.tuberculosis_H37Rv RPS-------------------------------------
MAB_1404|M.abscessus_ATCC_1997 LAQRMR----------------------------------
MSMEG_6230|M.smegmatis_MC2_155 ------------RR--------------------------
TH_1720|M.thermoresistible__bu PAVPARSADRTDRR--------------------------
Mflv_2361|M.gilvum_PYR-GCK ----------------------------------------
Mvan_4285|M.vanbaalenii_PYR-1 ARSDGSNAASALPRCDEASFSAGLSSAAVLAEPVGRNTGS