For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEVRAISGETPVEDHGYRPAPPGLRANMIFSADGAAAFGGRAGPLSCPADYQLLLALRAYADVVLVGAGT ARAESYGPVALRPDHRDQRRELGLGVQPPPLAVVSRSGRLPESIFAGPVPPILITSAQGAQECSEKRCQI VVSGDDAVDVAGAVHQLRSDGMERVLCEGGPTLLDELVTADLVDELCVTLAPRLAGSQPVGSGTPADIAA PAPMRLGRVLVDPDGYLFLTYRRA*
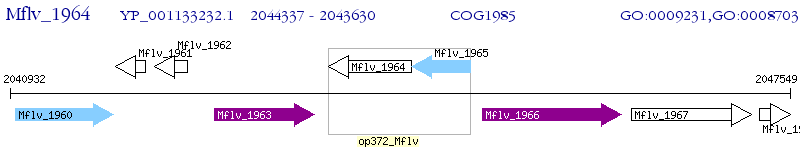
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1964 | - | - | 100% (235) | hypothetical protein Mflv_1964 |
| M. gilvum PYR-GCK | Mflv_3913 | - | 2e-33 | 41.48% (229) | hypothetical protein Mflv_3913 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2690 | ribD | 1e-29 | 37.02% (235) | hypothetical protein Mb2690 |
| M. tuberculosis H37Rv | Rv2671 | ribD | 1e-29 | 37.02% (235) | hypothetical protein Rv2671 |
| M. leprae Br4923 | MLBr_01340 | - | 1e-28 | 33.74% (246) | hypothetical protein MLBr_01340 |
| M. abscessus ATCC 19977 | MAB_2976 | - | 1e-34 | 43.72% (215) | hypothetical protein MAB_2976 |
| M. marinum M | MMAR_2045 | ribD | 3e-31 | 37.30% (244) | bifunctional enzyme riboflavin biosynthesis protein RibD |
| M. avium 104 | MAV_3050 | - | 5e-62 | 59.82% (219) | hypothetical protein MAV_3050 |
| M. smegmatis MC2 155 | MSMEG_2787 | - | 5e-27 | 38.57% (223) | hypothetical protein MSMEG_2787 |
| M. thermoresistible (build 8) | TH_1514 | ribD | 1e-12 | 45.56% (90) | POSSIBLE BIFUNCTIONAL ENZYME RIBOFLAVIN BIOSYNTHESIS PROTEIN |
| M. ulcerans Agy99 | MUL_3305 | ribD | 1e-29 | 36.07% (244) | hypothetical protein MUL_3305 |
| M. vanbaalenii PYR-1 | Mvan_4752 | - | 5e-88 | 69.66% (234) | hypothetical protein Mvan_4752 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2045|M.marinum_M MPD--------PTSGRSADAQLTLLGTDRQLTDEDLP-EFYGYPAKLSGT
MUL_3305|M.ulcerans_Agy99 MPD--------PTSGRSADAQLTLLGTDRQLTDEDLP-EFYGYPAKLSST
Mb2690|M.bovis_AF2122/97 MPD-------SGQLG-AADTPLRLLSSVHYLTDGELP-QLYDYPDD--GT
Rv2671|M.tuberculosis_H37Rv MPD-------SGQLG-AADTPLRLLSSVHYLTDGELP-QLYDYPDD--GT
MLBr_01340|M.leprae_Br4923 MSDFETGPAASTTAGRAAQTPLLMLTSGRYLDDDELS-QLYGYPSERAGV
MSMEG_2787|M.smegmatis_MC2_155 MSD---------DTAPARFTELTAGGRTVGSGDDELTTRFYAYPDDLRKC
TH_1514|M.thermoresistible__bu --------------------------------------------------
MAB_2976|M.abscessus_ATCC_1997 MADSDD----EQAIRGPAGTQLTQLTTGAPVDSGELI-DLYDFPKN-DKI
Mflv_1964|M.gilvum_PYR-GCK ------------------MTEVRAISGETPVEDHG----YRPAPPG----
Mvan_4752|M.vanbaalenii_PYR-1 ---------------------MRRLAEGAAVEDHG----YRPAPAG----
MAV_3050|M.avium_104 ------------------MNSPPRISAVPGIIELAPF--YADPPDG----
MMAR_2045|M.marinum_M WLRANFITSLDGGATVGGTSGGLAGPGDRAVFVLLRELADVILVGAGTVR
MUL_3305|M.ulcerans_Agy99 WLRANFITSLDGGATVGGTSGGLAGPGDRAVFVLLRELADVILVGAGTVR
Mb2690|M.bovis_AF2122/97 WLRANFISSLDGGATVDGTSGAMAGPGDRFVFNLLRELADVIVVGVGTVR
Rv2671|M.tuberculosis_H37Rv WLRANFISSLDGGATVDGTSGAMAGPGDRFVFNLLRELADVIVVGVGTVR
MLBr_01340|M.leprae_Br4923 WIRANFITSIDGGATADGRTGAMAGPGDRFVFNMLRELADVIVVGAGTVR
MSMEG_2787|M.smegmatis_MC2_155 RVRANMIVSLDGAATQDGKSGGLGGAGDRAVFNLMRETADVILMGASTVR
TH_1514|M.thermoresistible__bu --------------------------------------------------
MAB_2976|M.abscessus_ATCC_1997 YLRANMISSLDGAATVTGLSGGLGGDGDRAVFAAMRANADVILVGSGTVR
Mflv_1964|M.gilvum_PYR-GCK -LRANMIFSADGAAAFGGRAGPLSCPADYQLLLALRAYADVVLVGAGTAR
Mvan_4752|M.vanbaalenii_PYR-1 -LRANMIFSADGAAGFAGRAGPLSCPADHTLLLALRAYADVVLVGAGTAR
MAV_3050|M.avium_104 -VRANMIFSADGAAAFGGRAGPLSCRTDQRLLRILRGLADVVLVGAGTAR
MMAR_2045|M.marinum_M IEGYSGAQLGVAERQQRQARGQ-SEVPQLAIATNSGQLDRDLPVFTRTEV
MUL_3305|M.ulcerans_Agy99 IEGYSGAQRGVAERQQRQARGQ-SEVPQLAIATNSGQLDRDLPVFTRTEV
Mb2690|M.bovis_AF2122/97 IEGYSGVRMGVVQRQHRQARGQ-SEVPQLAIVTRSGRLDRDMAVFTRTEM
Rv2671|M.tuberculosis_H37Rv IEGYSGVRMGVVQRQHRQARGQ-SEVPQLAIVTRSGRLDRDMAVFTRTEM
MLBr_01340|M.leprae_Br4923 IENYSGAHLPVTKRQQRQARGQ-SEVPQLAIVTNSGRLDRDMAVFTRTEI
MSMEG_2787|M.smegmatis_MC2_155 IENYSGVQLSVPQRQARQRRGQ-AEVPPIAIVTASADLEPDAKIFTRTEV
TH_1514|M.thermoresistible__bu --------------------------------------------------
MAB_2976|M.abscessus_ATCC_1997 AERYHGAHLPVGLRQRRQSRGQ-DEVPTIAVVTASGAVDPSTPLFNESEV
Mflv_1964|M.gilvum_PYR-GCK AESYGPVALRPDHRDQRRELGLGVQPPPLAVVSRSGRLP--ESIFAG-PV
Mvan_4752|M.vanbaalenii_PYR-1 AENYGPVRLRAEHLAQRRELGLGEQPPAIAVVSRSGRLP--ESMFG--SV
MAV_3050|M.avium_104 AENYGPVRLTDAARARRLHEGR-AAPPPIAVVSRSGELP--ARLFSDPEQ
MMAR_2045|M.marinum_M PPLVLTAAAAVYDTRRRLTGLAEVVDCSGKDPHRVDEGLLLARLAGRGMR
MUL_3305|M.ulcerans_Agy99 PPLVLTAAAAVYDTRRRLTGLAEVVGCSGKDPHRVDEGLLLARLAGRGMR
Mb2690|M.bovis_AF2122/97 APLVLTTTAVADDTRQRLAGLAEVIACSGDDPGTVDEAVLVSQLAARGLR
Rv2671|M.tuberculosis_H37Rv APLVLTTTAVADDTRQRLAGLAEVIACSGDDPGTVDEAVLVSQLAARGLR
MLBr_01340|M.leprae_Br4923 APLVLTCTETAKKLRTRLADLAYVVSCSGDDPSKVDETVVLATLQTRGLR
MSMEG_2787|M.smegmatis_MC2_155 RPLILTTAHTVSDARRRLGGVAEVLDASGADRTRVDPARALQILAERKLF
TH_1514|M.thermoresistible__bu -----------------------VLDASGANPDSVDPAVVLRILSERGLW
MAB_2976|M.abscessus_ATCC_1997 APIVVTSASGAPHVASRVPGAQILVSGDGD---TVDLPAALATLHARGLS
Mflv_1964|M.gilvum_PYR-GCK PPILITSAQGAQECS---EKRCQIVVSGDD---AVDVAGAVHQLRSDGME
Mvan_4752|M.vanbaalenii_PYR-1 PPILVTSARGRQPRPSGVEKSCEVMVCGDE---QVDVAEAVRRLRAQGSH
MAV_3050|M.avium_104 PPILVTCARTAARPD---LDAQHVLVAGEE---TVDVARAVAMLREAGMH
:: . ** : *
MMAR_2045|M.marinum_M RILTEGGPTLFGSLIEQDLLDELCLTIAPVVVGGLPRRIASGPGQLQTR-
MUL_3305|M.ulcerans_Agy99 CILTEGGPTLFGSLIEQDLLDELCLTIAPVVVGGLPRRIASGPGQLQTR-
Mb2690|M.bovis_AF2122/97 RILTEGGPTLLGTFVERDVLDELCLTIAPYVVGGLARRIVTGPGQVLTR-
Rv2671|M.tuberculosis_H37Rv RILTEGGPTLLGTFVERDVLDELCLTIAPYVVGGLARRIVTGPGQVLTR-
MLBr_01340|M.leprae_Br4923 RVLTEGGPMLLGSFIQHGMLDELCLTIAPLIVGGRARRIATGPEQLLTR-
MSMEG_2787|M.smegmatis_MC2_155 RVLTEGGPSLLGLLIEHDLLDELCLTIAPVLVGGGAPRIAAGIGQARTR-
TH_1514|M.thermoresistible__bu RVLAEGGPTLLSQMLAAGLLDELCLTVAPVLVGGTGPRIVDGPHRVHQR-
MAB_2976|M.abscessus_ATCC_1997 RVLCEGGPALLGTLLSAHLVDELCLTIAPTTIGGAGLRITSGPAEVLTG-
Mflv_1964|M.gilvum_PYR-GCK RVLCEGGPTLLDELVTADLVDELCVTLAPRLAGSQPVG-SGTPADIAAPA
Mvan_4752|M.vanbaalenii_PYR-1 RILCEGGPTLLNELVEADLVDELCVTLAPRLAGSQPLG-AGRPARLTSPP
MAV_3050|M.avium_104 RILCEGGPTLLDELVEADALTELCVTLAPKLAASQPLGYRVHPARLTAPV
:* **** *:. :: : ***:*:** ..
MMAR_2045|M.marinum_M -MHCAHVITDDAGYLYTRYVRA-
MUL_3305|M.ulcerans_Agy99 -MHCAHVITDDAGYLYTRYVRA-
Mb2690|M.bovis_AF2122/97 -MRCAHVLTDDSGYLYTRYVKT-
Rv2671|M.tuberculosis_H37Rv -MRCAHVLTDDSGYLYTRYVKT-
MLBr_01340|M.leprae_Br4923 -MRCAHVLTDDAGYLYTRYVKA-
MSMEG_2787|M.smegmatis_MC2_155 -MRVSHLLTDDEGYLYTRYVRG-
TH_1514|M.thermoresistible__bu -MRRTHVLADDDGYLYTRYVKGD
MAB_2976|M.abscessus_ATCC_1997 -WRRVLLLADADGYLFTRYVPA-
Mflv_1964|M.gilvum_PYR-GCK PMRLGRVLVDPDGYLFLTYRRA-
Mvan_4752|M.vanbaalenii_PYR-1 GLRLAHILAGTDDYVYLKYVRP-
MAV_3050|M.avium_104 GLHLAHALVC-DDYLFLRYRR--
: :. .*:: *