For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPDPTSGRSADAQLTLLGTDRQLTDEDLPEFYGYPAKLSSTWLRANFITSLDGGATVGGTSGGLAGPGDR AVFVLLRELADVILVGAGTVRIEGYSGAQRGVAERQQRQARGQSEVPQLAIATNSGQLDRDLPVFTRTEV PPLVLTAAAAVYDTRRRLTGLAEVVGCSGKDPHRVDEGLLLARLAGRGMRCILTEGGPTLFGSLIEQDLL DELCLTIAPVVVGGLPRRIASGPGQLQTRMHCAHVITDDAGYLYTRYVRA
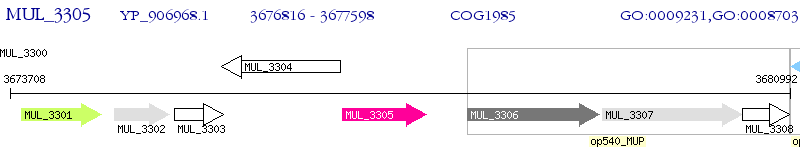
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3305 | ribD | - | 100% (260) | hypothetical protein MUL_3305 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2690 | ribD | 8e-99 | 68.73% (259) | hypothetical protein Mb2690 |
| M. gilvum PYR-GCK | Mflv_3913 | - | 2e-71 | 53.54% (254) | hypothetical protein Mflv_3913 |
| M. tuberculosis H37Rv | Rv2671 | ribD | 8e-99 | 68.73% (259) | hypothetical protein Rv2671 |
| M. leprae Br4923 | MLBr_01340 | - | 8e-94 | 65.23% (256) | hypothetical protein MLBr_01340 |
| M. abscessus ATCC 19977 | MAB_2976 | - | 1e-55 | 48.61% (251) | hypothetical protein MAB_2976 |
| M. marinum M | MMAR_2045 | ribD | 1e-144 | 98.46% (260) | bifunctional enzyme riboflavin biosynthesis protein RibD |
| M. avium 104 | MAV_3563 | - | 7e-97 | 70.68% (249) | hypothetical protein MAV_3563 |
| M. smegmatis MC2 155 | MSMEG_2787 | - | 4e-73 | 61.70% (235) | hypothetical protein MSMEG_2787 |
| M. thermoresistible (build 8) | TH_1514 | ribD | 6e-23 | 51.04% (96) | POSSIBLE BIFUNCTIONAL ENZYME RIBOFLAVIN BIOSYNTHESIS PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_2490 | - | 3e-74 | 55.91% (254) | hypothetical protein Mvan_2490 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3913|M.gilvum_PYR-GCK MSG------------SGDGTHFTLLGR----TDAVGVPDLAAHYPYPDGL
Mvan_2490|M.vanbaalenii_PYR-1 MSG------------DGDGTQFTLLGR----IEELGAEGVAAHYGYPDGL
MSMEG_2787|M.smegmatis_MC2_155 MSD------------DTAPARFTELTAGGRTVGSGDDELTTRFYAYPDDL
TH_1514|M.thermoresistible__bu --------------------------------------------------
MUL_3305|M.ulcerans_Agy99 MPD--------PTSGRSADAQLTLLGT----DRQLTDEDLPEFYGYPAKL
MMAR_2045|M.marinum_M MPD--------PTSGRSADAQLTLLGT----DRQLTDEDLPEFYGYPAKL
Mb2690|M.bovis_AF2122/97 MPD-------SGQLG-AADTPLRLLSS----VHYLTDGELPQLYDYPDD-
Rv2671|M.tuberculosis_H37Rv MPD-------SGQLG-AADTPLRLLSS----VHYLTDGELPQLYDYPDD-
MLBr_01340|M.leprae_Br4923 MSDFETGPAASTTAGRAAQTPLLMLTS----GRYLDDDELSQLYGYPSER
MAV_3563|M.avium_104 MAL--------------PETPLSLLGS----VREVDDGELPGLYDYPDRH
MAB_2976|M.abscessus_ATCC_1997 MAD----SDDEQAIRGPAGTQLTQLTT----GAPVDSGELIDLYDFPKN-
Mflv_3913|M.gilvum_PYR-GCK DSCWVRANMITSVDGGATSAGKSGGLGGDGDRALFSALRELADVIVVGAQ
Mvan_2490|M.vanbaalenii_PYR-1 QSCWVRANMIASVDGGATSGGKSGGLGGDGDRALFATLRDLADVIVVGAA
MSMEG_2787|M.smegmatis_MC2_155 RKCRVRANMIVSLDGAATQDGKSGGLGGAGDRAVFNLMRETADVILMGAS
TH_1514|M.thermoresistible__bu --------------------------------------------------
MUL_3305|M.ulcerans_Agy99 SSTWLRANFITSLDGGATVGGTSGGLAGPGDRAVFVLLRELADVILVGAG
MMAR_2045|M.marinum_M SGTWLRANFITSLDGGATVGGTSGGLAGPGDRAVFVLLRELADVILVGAG
Mb2690|M.bovis_AF2122/97 -GTWLRANFISSLDGGATVDGTSGAMAGPGDRFVFNLLRELADVIVVGVG
Rv2671|M.tuberculosis_H37Rv -GTWLRANFISSLDGGATVDGTSGAMAGPGDRFVFNLLRELADVIVVGVG
MLBr_01340|M.leprae_Br4923 AGVWIRANFITSIDGGATADGRTGAMAGPGDRFVFNMLRELADVIVVGAG
MAV_3563|M.avium_104 AGTWVRANFIASVDGGATADGSSGAMGGPGDRFVFNLLRELADVIVVGAG
MAB_2976|M.abscessus_ATCC_1997 DKIYLRANMISSLDGAATVTGLSGGLGGDGDRAVFAAMRANADVILVGSG
Mflv_3913|M.gilvum_PYR-GCK TVRSEDYSGVRLSASARLARQQRGQSEVPPIAVLTRSGRLDRDAKLFTRT
Mvan_2490|M.vanbaalenii_PYR-1 TVRVENYSGVQLGAAERLARQRRGQAEVPPVAVLTRSGLLDRDAKLFHLT
MSMEG_2787|M.smegmatis_MC2_155 TVRIENYSGVQLSVPQRQARQRRGQAEVPPIAIVTASADLEPDAKIFTRT
TH_1514|M.thermoresistible__bu --------------------------------------------------
MUL_3305|M.ulcerans_Agy99 TVRIEGYSGAQRGVAERQQRQARGQSEVPQLAIATNSGQLDRDLPVFTRT
MMAR_2045|M.marinum_M TVRIEGYSGAQLGVAERQQRQARGQSEVPQLAIATNSGQLDRDLPVFTRT
Mb2690|M.bovis_AF2122/97 TVRIEGYSGVRMGVVQRQHRQARGQSEVPQLAIVTRSGRLDRDMAVFTRT
Rv2671|M.tuberculosis_H37Rv TVRIEGYSGVRMGVVQRQHRQARGQSEVPQLAIVTRSGRLDRDMAVFTRT
MLBr_01340|M.leprae_Br4923 TVRIENYSGAHLPVTKRQQRQARGQSEVPQLAIVTNSGRLDRDMAVFTRT
MAV_3563|M.avium_104 TVRIEGYSGAQLGVAQRQQRQARGQSEVPPLGIVTKSGHLNRDMAVFTRT
MAB_2976|M.abscessus_ATCC_1997 TVRAERYHGAHLPVGLRQRRQSRGQDEVPTIAVVTASGAVDPSTPLFNES
Mflv_3913|M.gilvum_PYR-GCK DVPPLILTSTRALDDTRRLLGSVAEVLDASGAQPDSVDPAQALTLLAERG
Mvan_2490|M.vanbaalenii_PYR-1 DVPPLILTSSDAVEDTRQRLGAVAEVVDASGPAPDSVDLRTALDLLAERG
MSMEG_2787|M.smegmatis_MC2_155 EVRPLILTTAHTVSDARRRLGGVAEVLDASGADRTRVDPARALQILAERK
TH_1514|M.thermoresistible__bu -------------------------VLDASGANPDSVDPAVVLRILSERG
MUL_3305|M.ulcerans_Agy99 EVPPLVLTAAAAVYDTRRRLTGLAEVVGCSGKDPHRVDEGLLLARLAGRG
MMAR_2045|M.marinum_M EVPPLVLTAAAAVYDTRRRLTGLAEVVDCSGKDPHRVDEGLLLARLAGRG
Mb2690|M.bovis_AF2122/97 EMAPLVLTTTAVADDTRQRLAGLAEVIACSGDDPGTVDEAVLVSQLAARG
Rv2671|M.tuberculosis_H37Rv EMAPLVLTTTAVADDTRQRLAGLAEVIACSGDDPGTVDEAVLVSQLAARG
MLBr_01340|M.leprae_Br4923 EIAPLVLTCTETAKKLRTRLADLAYVVSCSGDDPSKVDETVVLATLQTRG
MAV_3563|M.avium_104 EVPPLVLTCAAAADQTRRLLSGLCEVLDCSAGDPERVDEATMLALLGERG
MAB_2976|M.abscessus_ATCC_1997 EVAPIVVTSASGAPHVASRVPGAQILVSGDG---DTVDLPAALATLHARG
:: .. ** : * *
Mflv_3913|M.gilvum_PYR-GCK LRRVLTEGGPGILGMFTDDDLVDELCVTVAPLLLGGGPGRIVTGDGEVRS
Mvan_2490|M.vanbaalenii_PYR-1 LVRVLTEGGPGILGMFTEQDLLDELCLTVAPLLLGGGSARIVTGLGEVRS
MSMEG_2787|M.smegmatis_MC2_155 LFRVLTEGGPSLLGLLIEHDLLDELCLTIAPVLVGGGAPRIAAGIGQART
TH_1514|M.thermoresistible__bu LWRVLAEGGPTLLSQMLAAGLLDELCLTVAPVLVGGTGPRIVDGPHRVHQ
MUL_3305|M.ulcerans_Agy99 MRCILTEGGPTLFGSLIEQDLLDELCLTIAPVVVGGLPRRIASGPGQLQT
MMAR_2045|M.marinum_M MRRILTEGGPTLFGSLIEQDLLDELCLTIAPVVVGGLPRRIASGPGQLQT
Mb2690|M.bovis_AF2122/97 LRRILTEGGPTLLGTFVERDVLDELCLTIAPYVVGGLARRIVTGPGQVLT
Rv2671|M.tuberculosis_H37Rv LRRILTEGGPTLLGTFVERDVLDELCLTIAPYVVGGLARRIVTGPGQVLT
MLBr_01340|M.leprae_Br4923 LRRVLTEGGPMLLGSFIQHGMLDELCLTIAPLIVGGRARRIATGPEQLLT
MAV_3563|M.avium_104 MRRILTEGGPTLLNSLLERQLLDELCLTIAPYLVGGHARRIATGPGQLLT
MAB_2976|M.abscessus_ATCC_1997 LSRVLCEGGPALLGTLLSAHLVDELCLTIAPTTIGGAGLRITSGPAEVLT
: :* **** ::. : ::****:*:** :** **. * .
Mflv_3913|M.gilvum_PYR-GCK GMVLGHALTDGDGYLYLRYLRDR-------
Mvan_2490|M.vanbaalenii_PYR-1 DLALLHALTDDRGFLYLRYVRDRLARDAER
MSMEG_2787|M.smegmatis_MC2_155 RMRVSHLLTDDEGYLYTRYVRG--------
TH_1514|M.thermoresistible__bu RMRRTHVLADDDGYLYTRYVKGD-------
MUL_3305|M.ulcerans_Agy99 RMHCAHVITDDAGYLYTRYVRA--------
MMAR_2045|M.marinum_M RMHCAHVITDDAGYLYTRYVRA--------
Mb2690|M.bovis_AF2122/97 RMRCAHVLTDDSGYLYTRYVKT--------
Rv2671|M.tuberculosis_H37Rv RMRCAHVLTDDSGYLYTRYVKT--------
MLBr_01340|M.leprae_Br4923 RMRCAHVLTDDAGYLYTRYVKA--------
MAV_3563|M.avium_104 RMRCAHVLTDDAGYLYTRYVRC--------
MAB_2976|M.abscessus_ATCC_1997 GWRRVLLLADADGYLFTRYVPA--------
::* *:*: **: