For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SALSRFVRRTPSLRQRVAFTSAIAGAIVVLIAGTVVWFGITDAWNERLDRRLDEAAGFVIPFLPRGLDEI PPSPNDQDVVITVRRDGGDAASNSTVVLPEMEPGYADTYLDGVRYRVRTVELATPQPMSVAVGATYDATI ADINNLHRRVLIICTLAIGAATLGGWVLAAFAVRPFKQLARQTSAIDTGDEEPDIDVRGATEAVEIAEAI NGLVQRVWEEQERTKAALTSARDFASVSAHELRTPLTAMRTNLEVLATLELSEDGRNEVLGDVMRTQTRI EATLGALERLAQGELSTEDDHVPVDITELLDRAAHDAMRVYPDLDVSLVPAPTVIIVGLPAGLRLAVDNA IANAVKHGGATRVQLSAVSSRDGVEIAIDDDGVGVPEAEREVVFDRFSRGSTASQSGSGLGLALVAQQAE LHGGTASLGSSPLGGARLLLRLPGPH*
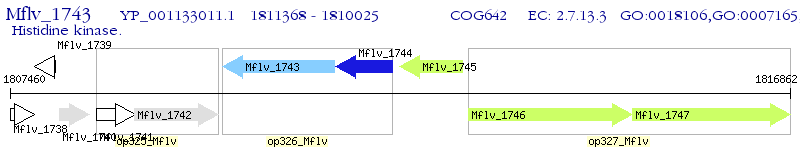
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1743 | - | - | 100% (447) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_4243 | - | e-130 | 56.78% (435) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_0460 | - | 3e-72 | 38.26% (447) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0926c | prrB | 0.0 | 76.46% (446) | two component sensor histidine kinase PRRB |
| M. tuberculosis H37Rv | Rv0902c | prrB | 0.0 | 76.46% (446) | two component sensor histidine kinase PRRB |
| M. leprae Br4923 | MLBr_02124 | - | 0.0 | 75.11% (446) | sensor histidine kinase |
| M. abscessus ATCC 19977 | MAB_0955c | - | 1e-130 | 56.78% (435) | sensor histidine kinase PrrB |
| M. marinum M | MMAR_4634 | prrB | 0.0 | 76.91% (446) | two-component sensor histidine kinase PrrB |
| M. avium 104 | MAV_1021 | - | 0.0 | 77.88% (443) | sensor-type histidine kinase PrrB |
| M. smegmatis MC2 155 | MSMEG_5663 | - | 0.0 | 79.15% (446) | sensor-type histidine kinase PrrB |
| M. thermoresistible (build 8) | TH_0454 | - | 0.0 | 78.03% (446) | sensor-type histidine kinase PrrB |
| M. ulcerans Agy99 | MUL_0249 | prrB | 0.0 | 76.68% (446) | two component sensor histidine kinase PrrB |
| M. vanbaalenii PYR-1 | Mvan_5010 | - | 0.0 | 85.20% (446) | integral membrane sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0926c|M.bovis_AF2122/97 ----------------------MNILSRIFARTP---SLRTRVVVATAIG
Rv0902c|M.tuberculosis_H37Rv ----------------------MNILSRIFARTP---SLRTRVVVATAIG
MMAR_4634|M.marinum_M -------------------MTVMNILSRIFARTP---SLRTRVVVATAIG
MUL_0249|M.ulcerans_Agy99 ----------------------MNILSRIFARTP---SLRTRVVVATAIG
MLBr_02124|M.leprae_Br4923 ----------------------MNILSRIFARTP---SLRTRVVVATAIG
MAV_1021|M.avium_104 ----------------------MKLLSRIVTRTP---SLRARVAVATAIG
Mflv_1743|M.gilvum_PYR-GCK ----------------------MSALSRFVRRTP---SLRQRVAFTSAIA
Mvan_5010|M.vanbaalenii_PYR-1 ----------------------MIALSRIFRRTP---SLRTRVAFATAIA
MSMEG_5663|M.smegmatis_MC2_155 ----------------------MSMLTRIFRRTP---SLRTRVAFATAIG
TH_0454|M.thermoresistible__bu ----------------------MNLVTRLFRRTP---SLRTRVAFATAIA
MAB_0955c|M.abscessus_ATCC_199 MSTSHENPPVMGRPSKHRSQRITDLSYRLLEKPMRIFSLRTLVAIA-ALG
*:. :. *** *..: *:.
Mb0926c|M.bovis_AF2122/97 AAIPVLIVGTVVWVGITNDRKERLDRRLDEAAGF-AIPFVPRGLDEIPRS
Rv0902c|M.tuberculosis_H37Rv AAIPVLIVGTVVWVGITNDRKERLDRRLDEAAGF-AIPFVPRGLDEIPRS
MMAR_4634|M.marinum_M AAIPVLIVGTVVWVGITNDRKERLDRRLDEAAGF-AIPFVPRGLDEIPRS
MUL_0249|M.ulcerans_Agy99 AAIPVLIVGTVVWVGITNDRKERLDRRLDEAAGF-AIPFVPRGLDEIPRS
MLBr_02124|M.leprae_Br4923 AAIPVLIVGTVVWVGITNDRKERLDRKLDEAAGF-AIPFVPRGLDEIPRS
MAV_1021|M.avium_104 TAIVVVIAGAVVWFGITSAWRERLDRRLDETAGF-AIPFLPRGLNEIPRS
Mflv_1743|M.gilvum_PYR-GCK GAIVVLIAGTVVWFGITDAWNERLDRRLDEAAGF-VIPFLPRGLDEIPPS
Mvan_5010|M.vanbaalenii_PYR-1 AAIVVGIVGTVVWVGITNDRQERLDRRLDEAAGF-AIPLLPRSLDEIPKS
MSMEG_5663|M.smegmatis_MC2_155 AAIVVVIVGTIVWIGITNDRKERLDRRLDEAAGF-AIPFLPRGLDQIPRS
TH_0454|M.thermoresistible__bu AAIVVGIVGVVVWVGITNDRKERLDRRLDEAAGF-ALPFVPRGLEQIPPS
MAB_0955c|M.abscessus_ATCC_199 VMVLVVTLGTWVWLGVEEDQYSQLDRRLDSVSSLGAVSNLLNSVISSDKK
: * *. **.*: . .:***:**..:.: .:. : ..: . .
Mb0926c|M.bovis_AF2122/97 PNDQD-ALITVR-RGNVIKSNSDITLPKLQDDYADTYVRGVRYRVRTVEI
Rv0902c|M.tuberculosis_H37Rv PNDQD-ALITVR-RGNVIKSNSDITLPKLQDDYADTYVRGVRYRVRTVEI
MMAR_4634|M.marinum_M PNDQD-ALITVR-RGDLVKSNSDITLPKLQKDYADTYIRGVRYRVRTVEI
MUL_0249|M.ulcerans_Agy99 PNDQD-ALITVR-RGDLVKSNSDITLPKLQEDYADTYIRGVRYRVRTVEI
MLBr_02124|M.leprae_Br4923 PNDQD-AIITVR-RGNLVKSNFDITLPKLTNDYADTYLRGVRYRVRTVEI
MAV_1021|M.avium_104 PKDQD-TVITVR-RDGQVKSNSDVTLPPMEEGYADTYINGVRYRVRTVEV
Mflv_1743|M.gilvum_PYR-GCK PNDQD-VVITVRRDGGDAASNSTVVLPEMEPGYADTYLDGVRYRVRTVEL
Mvan_5010|M.vanbaalenii_PYR-1 PNGQD-ALITVR-TGGQVTSNSNVVLPELEPGYADTYVDGVRYRVRTVAL
MSMEG_5663|M.smegmatis_MC2_155 PNDQD-AVITVR-QTGAVTSNSTVVLPELPAGYADTYIDGVRWRVRTVEI
TH_0454|M.thermoresistible__bu PNDQD-VVITVR-RGDEVRSNSEVVLPELDADYADTYIDGVRYRVRTVQT
MAB_0955c|M.abscessus_ATCC_199 LDTPDGLVLTARVAGLTKSIPTDTELPLLEPGYANTKINGVEYRVR--SF
. * ::*.* ** : .**:* : **.:***
Mb0926c|M.bovis_AF2122/97 PGPEPTSVAVGATYDATVAETNNLHRRVLLICTFAIGAAAVFAWLLAAFA
Rv0902c|M.tuberculosis_H37Rv PGPEPTSVAVGATYDATVAETNNLHRRVLLICTFAIGAAAVFAWLLAAFA
MMAR_4634|M.marinum_M PGPEPTSVAVGATYDATIAETNNLHRRVLLICTFAIGAATVFAWLLAVFA
MUL_0249|M.ulcerans_Agy99 PGPEPTSVAVGATYDATIAETNNLHRRVLLICTFAIGAATVFAWLLAVFA
MLBr_02124|M.leprae_Br4923 PAPEPTSIAVGATYDATVAETNNLHRRVLLICGFAIAAAAVFAWLLAAFA
MAV_1021|M.avium_104 RTPQPATVEVGATYDDTIAQTNNLHRRVILICALSIGAAAVFAWLLAAFA
Mflv_1743|M.gilvum_PYR-GCK ATPQPMSVAVGATYDATIADINNLHRRVLIICTLAIGAATLGGWVLAAFA
Mvan_5010|M.vanbaalenii_PYR-1 STPEPMSVAVGATYDATIADTNNLHRRVLIICTLAVGAATFGGWLLAAFA
MSMEG_5663|M.smegmatis_MC2_155 PAPGPMWVAVGATYDATIADTNNLHRRVLVICVFAVGAATVLGWVLAAFA
TH_0454|M.thermoresistible__bu PYPIRTTLAVGATYDDTIADTANLHRRVILICTLAIGAATIAGWLLAAFA
MAB_0955c|M.abscessus_ATCC_199 IAPGNIPVAVGAPVAETQRQIDRYHMRILAICGGVLIGTVIVGWVISLIV
* : ***. * : . * *:: ** : .:.. .*::: :.
Mb0926c|M.bovis_AF2122/97 VRPFKQLAEQTRSIDAGDEAPRVEVHGASEAIEIAEAMRGMLQRIWNEQN
Rv0902c|M.tuberculosis_H37Rv VRPFKQLAEQTRSIDAGDEAPRVEVHGASEAIEIAEAMRGMLQRIWNEQN
MMAR_4634|M.marinum_M VRPFKQLAEQTRSIDAGDEAPRVEVHGASEAIEIAEAMRGMLQRIWNEQI
MUL_0249|M.ulcerans_Agy99 VRPFKQLAEQTRSIDAGDEAPRVEVHGASEAIEIAEGMRGMLQRIWNEQI
MLBr_02124|M.leprae_Br4923 VRPFKQLAQQTRSVDAGGEAPRVEVHGATEAVEIAEAMRGMLQRIWNEQN
MAV_1021|M.avium_104 VRPFKRLAQQARSIDA-DERPQVAVRGASEAVEIAEAMRGMLQRIWKEQD
Mflv_1743|M.gilvum_PYR-GCK VRPFKQLARQTSAIDTGDEEPDIDVRGATEAVEIAEAINGLVQRVWEEQE
Mvan_5010|M.vanbaalenii_PYR-1 VRPFKRLAQQTRQIEAGGEAPDIDIRGATEAVEIADAIKGLVERVWEEQG
MSMEG_5663|M.smegmatis_MC2_155 VRPLKRLAEQTREIEAGDEAPDVEVRGATEAVEIAEAVNGMLKRIWKEQD
TH_0454|M.thermoresistible__bu VRPFKQLAAQARSIDAGDEVPDVEIRGATEAVEIADAVKGMLQRIWKEQD
MAB_0955c|M.abscessus_ATCC_199 VSPFRLLAQQARLINAQSNPDDVNVRGVREAVEISEAVEGMLERINTEQE
* *:: ** *: ::: .: : ::*. **:**::.:.*:::*: **
Mb0926c|M.bovis_AF2122/97 RTKEALASARDFAAVSSHELRTPLTAMRTNLEVLSTLDLPDDQRKEVLND
Rv0902c|M.tuberculosis_H37Rv RTKEALASARDFAAVSSHELRTPLTAMRTNLEVLSTLDLPDDQRKEVLND
MMAR_4634|M.marinum_M RTKEALASARDFAAVSSHELRTPLTAMRTNLEVLSTLDLPDDQRKEVLND
MUL_0249|M.ulcerans_Agy99 RTKEALASARDFAAVSSHELRTPLTAMRTNLEVLSTLDLPDDQRKEVLND
MLBr_02124|M.leprae_Br4923 RTKEALASARDFAAVSSHELRTPLTAMRTNLEVLATLDLADDQRKEVLGD
MAV_1021|M.avium_104 RTKEALASARDFSAVTAHELRSPLTAMRTNLEVLSTLDMPDEQRKEVLGD
Mflv_1743|M.gilvum_PYR-GCK RTKAALTSARDFASVSAHELRTPLTAMRTNLEVLATLELSEDGRNEVLGD
Mvan_5010|M.vanbaalenii_PYR-1 RTKAALASARDFASVSAHELRTPLTAMRTNVEVLATLDLSEEGRKEVVND
MSMEG_5663|M.smegmatis_MC2_155 RTKSALASARDFASVSAHELRTPLTAMRTNLEVLSTLDLPDEQRKEVVND
TH_0454|M.thermoresistible__bu RTKAALASARDFASAAAHELRTPLTAMRTNLEVLSTLDMPEEQRKEVLAD
MAB_0955c|M.abscessus_ATCC_199 KTKAALESARDFAAVASHELRTPLTAMRTNLEVLSTLNLSDDQREEIIEE
:** ** *****::.::****:********:***:**::.:: *:*:: :
Mb0926c|M.bovis_AF2122/97 VIRTQSRIEATLSALERLAQGELSTSDDHVPVDITDLLDRAAHDAARIYP
Rv0902c|M.tuberculosis_H37Rv VIRTQSRIEATLSALERLAQGELSTSDDHVPVDITDLLDRAAHDAARIYP
MMAR_4634|M.marinum_M VIRTQSRIEATLGALERLAQGELSTSEDHVPVDITELLDRAAHDAMRIYP
MUL_0249|M.ulcerans_Agy99 VIRTQSRIEATLGALERLAQGELSTSEDHVPVDITELLDRAAHDAMRIYP
MLBr_02124|M.leprae_Br4923 VIRTQSRIEATLSALERLAQGELSTSDDHVPVDITELLDRAAHDATRSYP
MAV_1021|M.avium_104 VIRTQSRIEATLTALERLAQGELSTSEDHVPVDITELLDRAAHDAMRIYP
Mflv_1743|M.gilvum_PYR-GCK VMRTQTRIEATLGALERLAQGELSTEDDHVPVDITELLDRAAHDAMRVYP
Mvan_5010|M.vanbaalenii_PYR-1 VIRTQSRIEATLGALERLAQGELSTEDDHVPVDITELLDRAAHDAMRVYP
MSMEG_5663|M.smegmatis_MC2_155 VIRTQSRIEATLGALERLAQGELSTTDDHVTVDITELLDRAAHDAMRIYP
TH_0454|M.thermoresistible__bu VIRTQSRIEATLSALERLAQGELSTSDDHVPVDITELLDRAAHEAMRIYP
MAB_0955c|M.abscessus_ATCC_199 TIRTQSRVEATLSALERLAQGQLTTEDDHVPVDITELLDRAAHDADRSYP
.:***:*:**** ********:*:* :***.****:*******:* * **
Mb0926c|M.bovis_AF2122/97 DLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGATLVQLSAVSSRA
Rv0902c|M.tuberculosis_H37Rv DLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGATLVQLSAVSSRA
MMAR_4634|M.marinum_M DLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGSTRVQLSAVSSRA
MUL_0249|M.ulcerans_Agy99 DLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGSTRVQLSAVSSRA
MLBr_02124|M.leprae_Br4923 ELKVSLVPSPTCIIVGLPAGLRLAVDNAVANAVKHGGATRVQLSAVSSRA
MAV_1021|M.avium_104 GLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGATRVQLSAVSSRA
Mflv_1743|M.gilvum_PYR-GCK DLDVSLVPAPTVIIVGLPAGLRLAVDNAIANAVKHGGATRVQLSAVSSRD
Mvan_5010|M.vanbaalenii_PYR-1 DLEVSLVPAPTVIIVGLPAGLRLAVDNAIANAVKHGGASRVQLSAVSSRE
MSMEG_5663|M.smegmatis_MC2_155 DLEVSLAPSPTVIIVGLPTGLRLSVDNAIANAVKHGGATRVQLSAVSSRE
TH_0454|M.thermoresistible__bu DLEVSLVPAPTVIIVGLPAGLRLAVDNAIANAVKHGGATRVQLSAVSSRE
MAB_0955c|M.abscessus_ATCC_199 NLEISLVPSSTILMLGLPAGLRLVIDNAIKNAVKHGGASKIQLSATSTVH
*.:**.*:.* :::***:**** :***: *******:: :****.*:
Mb0926c|M.bovis_AF2122/97 GVEIAIDDNGSGVPEGERQVVFERFSRGSTASHSGSGLGLALVAQQAQLH
Rv0902c|M.tuberculosis_H37Rv GVEIAIDDNGSGVPEGERQVVFERFSRGSTASHSGSGLGLALVAQQAQLH
MMAR_4634|M.marinum_M GVEIAVDDNGSGVPEAERQMVFERFSRGSTASHSGSGLGLALVAQQAHLH
MUL_0249|M.ulcerans_Agy99 GVEIAVDDNGSGVPEAERQMVFERFSRGSTASHSGSGLGLALVAQQAHLH
MLBr_02124|M.leprae_Br4923 GVEIAVDDNGSGVPEDERQVVFERFSRGSTASHSGSGLGLALVAQQAQLH
MAV_1021|M.avium_104 GVEIAVDDNGSGVPEDERQVVFDRFSRGSTASQSGSGLGLALVAQQAQLH
Mflv_1743|M.gilvum_PYR-GCK GVEIAIDDDGVGVPEAEREVVFDRFSRGSTASQSGSGLGLALVAQQAELH
Mvan_5010|M.vanbaalenii_PYR-1 GVEIAIDDDGVGVPEEERTVVFDRFSRGSTASHSGSGLGLALVAQQAELH
MSMEG_5663|M.smegmatis_MC2_155 GVEIAIDDNGVGVPEGERALVFDRFARGSTASRSGSGLGLALVAQQAELH
TH_0454|M.thermoresistible__bu GVEIAIDDNGTGVPEEERERVFDRFTRGSTASHSGSGLGLALVAQQAEIH
MAB_0955c|M.abscessus_ATCC_199 GVEIAVDDNGSGIPEAERVQVFERFSRGSTASHSGSGLGLALVAQQAELH
*****:**:* *:** ** **:**:******:**************.:*
Mb0926c|M.bovis_AF2122/97 GGTASLENSPLGGARLVLRLPGPS
Rv0902c|M.tuberculosis_H37Rv GGTASLENSPLGGARLVLRLPGPS
MMAR_4634|M.marinum_M GGTASLQNSPLGGARLLLKLPGPS
MUL_0249|M.ulcerans_Agy99 GGTASLQNSPLGGARLLLKLPGPS
MLBr_02124|M.leprae_Br4923 GGTASLETSPLGGARLLLRISAPS
MAV_1021|M.avium_104 GGTAALENSPLGGARLVLRLPGPS
Mflv_1743|M.gilvum_PYR-GCK GGTASLGSSPLGGARLLLRLPGPH
Mvan_5010|M.vanbaalenii_PYR-1 GGTASLEASPLGGARLLLRLPGPR
MSMEG_5663|M.smegmatis_MC2_155 GGTATLAESPLGGARLLLRLPGPR
TH_0454|M.thermoresistible__bu GGTASLEASPLGGARLVLRLPGPR
MAB_0955c|M.abscessus_ATCC_199 GGTAALESSPLGGARLVLRLPGH-
****:* ********:*::..