For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLKVTAAVTPKGERRRYALVSAAADLLCEGGFDAVRHRAVARRAGLPLASTTYYFSSLDDLIANAVEHVG SRETEQLRHRVAALSRRRRGAESTADILVDLLVGDNPERVTEQMISRYERYIACARQPGLRDIQRRILRQ RSDAVVEVVERSGRSVRAELVTALVCAVDGAVVAALVGDGDGPRATARATLIDVIDVLAPFN
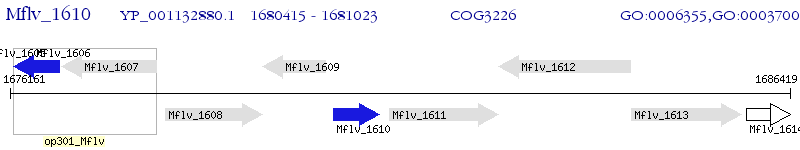
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1610 | - | - | 100% (202) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3251 | - | 2e-07 | 31.68% (101) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4816 | - | 3e-06 | 36.36% (88) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0798 | - | 1e-73 | 72.73% (198) | hypothetical protein Mb0798 |
| M. tuberculosis H37Rv | Rv0775 | - | 1e-73 | 72.73% (198) | hypothetical protein Rv0775 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0686 | - | 3e-60 | 60.94% (192) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4918 | - | 3e-75 | 75.25% (198) | hypothetical protein MMAR_4918 |
| M. avium 104 | MAV_0721 | - | 1e-63 | 67.39% (184) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5850 | - | 3e-92 | 87.37% (198) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1265 | - | 2e-85 | 81.54% (195) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0488 | - | 4e-71 | 74.21% (190) | hypothetical protein MUL_0488 |
| M. vanbaalenii PYR-1 | Mvan_5148 | - | 1e-102 | 94.97% (199) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1610|M.gilvum_PYR-GCK -------------MLKVTAAVTPKGERRRYALVSAAADLLCEGGFDAVRH
Mvan_5148|M.vanbaalenii_PYR-1 ----------------MTAAVTPKGERRRYALVSAAADLLCEGGFDAVRH
MSMEG_5850|M.smegmatis_MC2_155 ----------------MTAAVTPKGERRRYALIRAAAELLCEGGFDAVRH
TH_1265|M.thermoresistible__bu --------------------VTPKGERRRDALISAAAELLCEGGFDAVRH
MMAR_4918|M.marinum_M --------------MGVTAAVTPKGERRRYALVSAAAELLGEGGFEAVRH
MUL_0488|M.ulcerans_Agy99 ----------------MTAAVTPKGERRRYALVSAAAELLGEGGFEAVRH
Mb0798|M.bovis_AF2122/97 --------------MGVTAAVTPKGERRRYALVSAAAELLGEGGFEAVRH
Rv0775|M.tuberculosis_H37Rv --------------MGVTAAVTPKGERRRYALVSAAAELLGEGGFEAVRH
MAV_0721|M.avium_104 --------------------------------MSAAAELLAEGGFEAVRH
MAB_0686|M.abscessus_ATCC_1997 MTVRQHAEVFPDGNAAHRPQATPKGERRRRALISAAADLLRDNGIDAVRH
: ***:** :.*::****
Mflv_1610|M.gilvum_PYR-GCK RAVARRAGLPLASTTYYFSSLDDLIANAVEHVGSRETEQLRHRVAALSRR
Mvan_5148|M.vanbaalenii_PYR-1 RAVARRAGLPLASTTYYFSSLDDLIAKAVEHVGTRESEQLRTRVETLSRR
MSMEG_5850|M.smegmatis_MC2_155 RAVARRAGLPLASTTYYFSSLDDLIAKAVEYIGMREANQLRESVASLSRR
TH_1265|M.thermoresistible__bu RAVARRAGLPLASTTYYFSSLDDLIVQAVTHIGARETRQLEQRVAAVARR
MMAR_4918|M.marinum_M RAVARRAGLPLASTTYYFSSLDDLIARAVEHVGMIEVAQLRARVNALSRR
MUL_0488|M.ulcerans_Agy99 RAVARRAGLPLASTTYYFSSLDDLVARAVEHVGMIEVAQLRARVNALSRR
Mb0798|M.bovis_AF2122/97 RAVARRAGLPLASTTYYFSSLDDLIARAVEHIGMIEVAQLRARVSALSRR
Rv0775|M.tuberculosis_H37Rv RAVARRAGLPLASTTYYFSSLDDLIARAVEHIGMIEVAQLRARVSALSRR
MAV_0721|M.avium_104 RAVARRAGLPLASTTYYFSSLDDLIARAVEHIAMIEVAQLRSRVSALSRR
MAB_0686|M.abscessus_ATCC_1997 RAVAQRAGLPLASTTYYFSSLEDLIARAVEHAGTHELEQIQERLSLVQYR
****:****************:**:..** : . * *:. : : *
Mflv_1610|M.gilvum_PYR-GCK RRGAESTADILVDLLVGDNP-ERVTEQMISRYERYIACARQPGLRDIQRR
Mvan_5148|M.vanbaalenii_PYR-1 RRGAESTADIIVDLLVGDNP-ERVTEQIISRYERYIACARQPGLRDIQRR
MSMEG_5850|M.smegmatis_MC2_155 RRGAESTAEILVDLLVGDEPGARVTEELISRYERYIACARQPGLRDIQRR
TH_1265|M.thermoresistible__bu RRGAETTADVLVDVLVGTASGITATEELISRYERYIACARQPALRDIQRQ
MMAR_4918|M.marinum_M RRGPETTADVLVDLLVGDVSSPGLAEQLISRYERNIACTRLPALRESMRR
MUL_0488|M.ulcerans_Agy99 RRGPETTADVLVDLLVGDVSSPGLAEQLISRYERNIACTRLPALRESMRR
Mb0798|M.bovis_AF2122/97 RRGPETTAVVLVDLLVGEMSSPGLAEQLISRYERHIACTRLPDLRESMRR
Rv0775|M.tuberculosis_H37Rv RRGPETTAVVLVDLLVGEMSSPGLAEQLISRYERHIACTRLPDLRESMRR
MAV_0721|M.avium_104 RRGPETIAEVLADLLVGDVSGPGLTEQLISRYERHIACTRLPALRETMRR
MAB_0686|M.abscessus_ATCC_1997 RRGAAATAEAVVELLFG-TPGFGGGEQLVSRYERELACSRHPELREVQQR
***. : * :.::*.* . *:::***** :**:* * **: ::
Mflv_1610|M.gilvum_PYR-GCK ILRQRSDAVVEVVERSGRSVRAELVTALVCAVDGAVVAALVGDGDGPRAT
Mvan_5148|M.vanbaalenii_PYR-1 ILRQRSDAVVEVVERSGRSVRTELVTALVCAVDGAVVAALVGDGEGPRAT
MSMEG_5850|M.smegmatis_MC2_155 ILQQRTDAVVEAVERSGRSVRAELVTALVCAVDGAVVASLVDEGDGPRAS
TH_1265|M.thermoresistible__bu IRKQRTDAVVKVVERSGRAVRPELVSALVCAVDGAVVAALVGDGDGPRAT
MMAR_4918|M.marinum_M SLRQRAEAVAEAIERSGRSVRIELVCTLICAVDGSVVSALVE-GRDPRAA
MUL_0488|M.ulcerans_Agy99 SLRQRAEAVAEAIERSGRSVRIELVCTLICAVDGSVVSALVE-GRDPRAA
Mb0798|M.bovis_AF2122/97 SLRQRAEAVAEAIERSGRSAQIELVCTLICAVDGSVVSALVE-GRDPRAA
Rv0775|M.tuberculosis_H37Rv SLRQRAEAVAEAIERSGRSAQIELVCTLICAVDGSVVSALVE-GRDPRAA
MAV_0721|M.avium_104 SLRQRAEAVAEAIERSGRSVHIDLVCTLICAVDGSVVSALVE-GRDPRAA
MAB_0686|M.abscessus_ATCC_1997 LREQRDDAVAQVVRRSGRSIDNDHVAVLISTVDGAMVGALTEAAASPTAT
.** :**.:.:.****: : * .*:.:***::*.:*. . .* *:
Mflv_1610|M.gilvum_PYR-GCK ARATLIDVIDVLAPFN--------------------
Mvan_5148|M.vanbaalenii_PYR-1 ARATLIDVIDVLAPFN--------------------
MSMEG_5850|M.smegmatis_MC2_155 ARATLIDVIDVLAPVDDRAVRV--------------
TH_1265|M.thermoresistible__bu ARSTLIDVIDVLAPYA--------------------
MMAR_4918|M.marinum_M ALATVVDLIDVLAPVDQRPVQV--------------
MUL_0488|M.ulcerans_Agy99 ALATVV------------------------------
Mb0798|M.bovis_AF2122/97 ALATVVDLIDVLAPVDQRPVPF--------------
Rv0775|M.tuberculosis_H37Rv ALATVVDLIDVLAPVDQRPVPF--------------
MAV_0721|M.avium_104 AQGAVVDLIEVLAPIDQRPVQI--------------
MAB_0686|M.abscessus_ATCC_1997 ATAMLVDVIDVLAPLEAEQLAESVPADEHRGRRAWI
* . ::