For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGSEPEGARPAPEAPSTARQTAWNYLVFALSKSSTLLMTIVVARILSPTEFGIFALAILVVNLFDYVKD LGVGSALVQNRRDWRVLAPTGLTLSVLFGLFASGVLAATAGVAARALGEPELTELIRVLAVGLAISALST IPAAWLRRSMNFSARLLPEFAGAVAKTGLTIGLAAAGYGVWSLAYGQLAASVIMTILYWRVARVRPRFAF DGPVCRELVRFGVPVTAVTLLAYAIYNVDYLAVGERLGATELGLYSMAYRIPELLVLSLCVVISEVLFSA LSGLQDDRDALTGHYLQALTVVAALTIPIGIGLAVAAQPAVGTLYGSAYAESAPILSVLAIYATVYSASF HAGDVFKAIGRPGILTAINAGKLAVLVGPIWWAAGHSALAVAVALLSVEGLHFLVRMTVLCRLTRTSWSS IAGALCRPAAAAVPMGLVLVGVGHVIDGLADPLELALMVVVGLCVYVLILRFTAAQLVKAAFEMVRARLS RSAATPATTKEDL
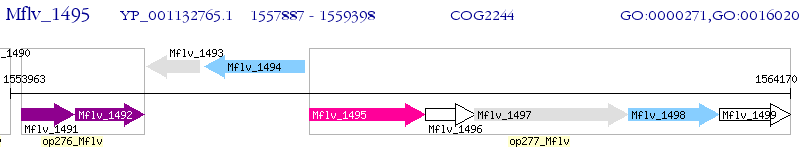
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1495 | - | - | 100% (503) | polysaccharide biosynthesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1017c | - | 8e-05 | 22.40% (433) | MOP superfamily O-antigen transporter |
| M. marinum M | MMAR_4504 | - | 1e-12 | 22.20% (473) | hypothetical protein MMAR_4504 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5968 | - | 7e-33 | 26.18% (489) | polysaccharide biosynthesis protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4675 | - | 9e-14 | 22.85% (477) | hypothetical protein MUL_4675 |
| M. vanbaalenii PYR-1 | Mvan_5251 | - | 0.0 | 86.04% (487) | polysaccharide biosynthesis protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1495|M.gilvum_PYR-GCK --------------MTGSEPEGARPAPEAPS-TARQTAWNYLVFALSKSS
Mvan_5251|M.vanbaalenii_PYR-1 ----------------------------MPS-TGRQTAWNYLIFALSKSS
MMAR_4504|M.marinum_M -------------MTEPTVPSSVVPSVPVAR-LAQAFSIQLICRILGMLA
MUL_4675|M.ulcerans_Agy99 -------------MTEPTAPSPVVPSVPVAR-LAQAFSIQLICRILGMLA
MSMEG_5968|M.smegmatis_MC2_155 MSSPGGRSDKSEALDPEDLPGAEAPVEPLGRSAAAGTLWLTAAQWLMRLT
MAB_1017c|M.abscessus_ATCC_199 ---------------MSNEPRSSTTLPRALR-LARDFGAVVFGKYGQYLV
.
Mflv_1495|M.gilvum_PYR-GCK TLLMTIVVARILSPTEFGIFALAILVVNLFDYVKDLGVGSALVQNRRDWR
Mvan_5251|M.vanbaalenii_PYR-1 TLLMTIVVARILSPTEFGIFALAVLVVNLFDYVKDLGVAAALVQNRREWR
MMAR_4504|M.marinum_M SVVSVMLTARYLGPGRYGQLTIAVAFIGMWASLVDLGIGRVIVRRVASSP
MUL_4675|M.ulcerans_Agy99 SVVSVMLTARYLGPGRYGQLTVAVAFIGMWASLVDLGIGRVIVRRVTSSP
MSMEG_5968|M.smegmatis_MC2_155 GLATIAILTRLLTPEDFGVVAAAMTVTPFLLLLADLGLSTYVLQVKRADQ
MAB_1017c|M.abscessus_ATCC_199 TFATVPLLARVLGPAGMGLLAVGMAAYFFGSIVVDLGMTPFLAARVPELR
. : :* * * * .: .: : : ***: :
Mflv_1495|M.gilvum_PYR-GCK -VLAPTGLTLSVLFGLFASGVLAATAGVAARALGEPELTELIRVLAVGLA
Mvan_5251|M.vanbaalenii_PYR-1 -ILAPTGLTLSVLFGLGASGVLAATAGVAARALGQPELTGLIRILAVGLA
MMAR_4504|M.marinum_M DDLERLVRIYNGLSLLYCVPLTLVAAGSGLLVYQDRDVRAMLVVLSCQLL
MUL_4675|M.ulcerans_Agy99 DDLERLVRIYNGLSLVYCIPLTLVAAGSGLLVYHDHEVRAMLVVLSCQLL
MSMEG_5968|M.smegmatis_MC2_155 -RLLSTAFWYSLTMAIVLMAAVAAFAPVIASLFDLPKAAPVLRGCSLAAG
MAB_1017c|M.abscessus_ATCC_199 NTPAELQQVRTDYLTVRLMTLGVLGAALGAGWIAG--VPPYVHMILLGLF
. : * :
Mflv_1495|M.gilvum_PYR-GCK ISALSTIPAAWLRRSMN-FSARLLPEFAGAVAKTGLTIGLAAAGYGVWSL
Mvan_5251|M.vanbaalenii_PYR-1 ISALSTIPAAWLRRSMN-FSARLMPEFAGAATKTGLTIGLAAAGFGVSSL
MMAR_4504|M.marinum_M IVTMLTRLEPVFLVTVR-FTAVAVSDVVGRLATLGVISFLVASRADVIWF
MUL_4675|M.ulcerans_Agy99 IVAMLTRLEPVFLVTVR-FTAVAVSDVVGRLATLGVISFLVASRPDVIWF
MSMEG_5968|M.smegmatis_MC2_155 FIILCSVPAALLRREMR-FRALSLQWLASAVIAQVVAVVLAFRGAGAWAL
MAB_1017c|M.abscessus_ATCC_199 TGGFWAASDDWLLIGQGRFIASAIYQGSGRITYLLLLVVLLPHSPHAATA
: : : * * : . : * .
Mflv_1495|M.gilvum_PYR-GCK AYGQLAASVIMTILYWRVARVRPRFA--FDGPVCRELVRFGVPVTAVTLL
Mvan_5251|M.vanbaalenii_PYR-1 AYGQLAAVVVMTVLYWRVAHIRPSLG--FDSRVCRELIRFGLPVTAVTLL
MMAR_4504|M.marinum_M AVPQLIQTGLQLLIQSRAASRRVSLRPILALREAADLLRESLFPMGVTVI
MUL_4675|M.ulcerans_Agy99 AVPQLIQTALQLLIQSRAASQRVRLRPILALREAADLLRESLFPMGVTVI
MSMEG_5968|M.smegmatis_MC2_155 VSQLVIAEAISCVLVWQAARWRPTLQ--FSKSEFFDMAKFGNKVIAADVI
MAB_1017c|M.abscessus_ATCC_199 ILCLLGSSILTVAGTWWDSLR--TFGSLCRPYAPGQILRSSWPIVTSRLL
: : : : :: : . ::
Mflv_1495|M.gilvum_PYR-GCK AYAIYNVDYLAVGERLGATELGLYSMAYRIPELLVLSLCVVISEVLFSAL
Mvan_5251|M.vanbaalenii_PYR-1 AYAIYNVDYLAIGERLGATELGLYTMAYRIPELLVLSLCVVVSEVLFSAL
MMAR_4504|M.marinum_M TVLYWRVDGVILSLLNTHSEVGVYGLASSIAINTLVLSVFFLKSTLSTAT
MUL_4675|M.ulcerans_Agy99 TVLYWRVDGVILSLLNTHSEVGVYGLASSIALNTLVLSVFFLKSTLSTAT
MSMEG_5968|M.smegmatis_MC2_155 VAMRPAAEAAIISTVLGPAALGFLSIAQRLVQVTQDLGGKALVPVSTVVF
MAB_1017c|M.abscessus_ATCC_199 TTGYGQGAPAIYSAMLDAVSLGLFSASDRLVRAMQSLLDMIGLVLLPRMA
. . :*. : :
Mflv_1495|M.gilvum_PYR-GCK SGLQDDRDALTGHYLQALTVVAALTIPIGIGLAVAAQPAVGTLYGSAYAE
Mvan_5251|M.vanbaalenii_PYR-1 SGLQDDRDALTGHYMQALTVVAALTFPIGVGLAVAAQPLVGTLYGVDYAG
MMAR_4504|M.marinum_M ELYSRDLAAFAGFMRRSVELMYFLAVPIAVVGVLVAQPLIRLLGSQAFVS
MUL_4675|M.ulcerans_Agy99 ELYSRDLAAFAGFIRRSVELMYFLVVPIAVVGVLVAQPLIRLLSSQAFVS
MSMEG_5968|M.smegmatis_MC2_155 AKVRESPERLQSAYLKALGIAYAAVSPLMTVVVVGGTLVVPLLFGQGWDH
MAB_1017c|M.abscessus_ATCC_199 K--RSGHEHFWRSGFQGLRGAVAVAVLAATLLWVLATPIVHLIFGSEFLG
. : :.: . : . : : . :
Mflv_1495|M.gilvum_PYR-GCK S-APILSVLAIYATVYSASFHAGDVFKAIGRPGILTAINAGKLAVLVGPI
Mvan_5251|M.vanbaalenii_PYR-1 S-APILSVLAIYTALYSASFHAGDVFKAIGRPGILTAINAGKLAVLVVPI
MMAR_4504|M.marinum_M RGTPTLALLFIAVGLRFIATTLGEGLFASHSQRFWFWLSVATLALNIVLN
MUL_4675|M.ulcerans_Agy99 RGTPTLALLFIAVGLRFIATTLGEGLFASHSQRFWFWLSVATLTLNIVLN
MSMEG_5968|M.smegmatis_MC2_155 S-VPVAQALSLAAILTLGAIIDQRLHYGIGRPGRWLVY-AGCIGAVEIAV
MAB_1017c|M.abscessus_ATCC_199 AVGLLRAELLILPASTISSYISTALLPVRQDTHGVLIAAVVGTTAAMAGL
* : : .
Mflv_1495|M.gilvum_PYR-GCK WWAAG-HSALAVAVALLSVEGLHFLVRMTVLCRLTR---TSWSSIAGALC
Mvan_5251|M.vanbaalenii_PYR-1 WWAAG-HSALMVAFALLGVEALHFLVRMTVLHRLMG---TSWLSIAGALC
MMAR_4504|M.marinum_M VTLDGRFGAVGAGIALVCTELFNMTIASWWLHRQCDY--RTPVLFLLRLS
MUL_4675|M.ulcerans_Agy99 LALDGRLGAVGAGIALVCTELFNMTIASWWLHRQCGY--HTPVLFLLRLS
MSMEG_5968|M.smegmatis_MC2_155 TALVAHNGLNWVAIGFVSVAFVATVIRWVLVGRLLDIPPRTLARVFAAAS
MAB_1017c|M.abscessus_ATCC_199 LAASWTHSVWALTGGVIGAEFCVALWYLARVRHLSGR--ERGAAATAETA
. ..: . : : .
Mflv_1495|M.gilvum_PYR-GCK RPAAAAVPMGLVLVGVGHVIDGLADPLELALMVVVGLCVYVLILRFTAAQ
Mvan_5251|M.vanbaalenii_PYR-1 RPAAAAVPMGLVLLGLARVTDGLADPLELVVLAFVGLCVYVLILRFTAAQ
MMAR_4504|M.marinum_M VPTAASVAMVLLLAG------------HHVIVVLAAAAVAYLAINAAIGP
MUL_4675|M.ulcerans_Agy99 VPTAASVAVVLPLAG------------HHVIVVLAAAAVAYLAINAAIGP
MSMEG_5968|M.smegmatis_MC2_155 VAVAGSAGAGLLVLSLTG---GWAPAWSLAAVMVAVGVVHIGIVRVVSPD
MAB_1017c|M.abscessus_ATCC_199 QEEAAR--------------------------------------------
*.
Mflv_1495|M.gilvum_PYR-GCK LVKAAFEMVRARLSRSAATPATTKEDL-
Mvan_5251|M.vanbaalenii_PYR-1 LVKAAFGMVRQRLSRSTAASATTAKEGP
MMAR_4504|M.marinum_M LTWSTMASLRPNSQPKQLTP--------
MUL_4675|M.ulcerans_Agy99 LTWSTMASLRPNAQPEQLTP--------
MSMEG_5968|M.smegmatis_MC2_155 VYRTVRSLVHRAPELDESL---------
MAB_1017c|M.abscessus_ATCC_199 ----------------------------