For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSPGGRSDKSEALDPEDLPGAEAPVEPLGRSAAAGTLWLTAAQWLMRLTGLATIAILTRLLTPEDFGVVA AAMTVTPFLLLLADLGLSTYVLQVKRADQRLLSTAFWYSLTMAIVLMAAVAAFAPVIASLFDLPKAAPVL RGCSLAAGFIILCSVPAALLRREMRFRALSLQWLASAVIAQVVAVVLAFRGAGAWALVSQLVIAEAISCV LVWQAARWRPTLQFSKSEFFDMAKFGNKVIAADVIVAMRPAAEAAIISTVLGPAALGFLSIAQRLVQVTQ DLGGKALVPVSTVVFAKVRESPERLQSAYLKALGIAYAAVSPLMTVVVVGGTLVVPLLFGQGWDHSVPVA QALSLAAILTLGAIIDQRLHYGIGRPGRWLVYAGCIGAVEIAVTALVAHNGLNWVAIGFVSVAFVATVIR WVLVGRLLDIPPRTLARVFAAASVAVAGSAGAGLLVLSLTGGWAPAWSLAAVMVAVGVVHIGIVRVVSPD VYRTVRSLVHRAPELDESL*
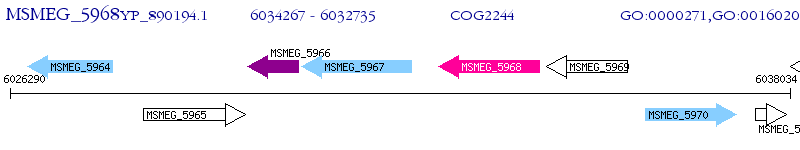
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5968 | - | - | 100% (510) | polysaccharide biosynthesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1495 | - | 6e-33 | 26.18% (489) | polysaccharide biosynthesis protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5251 | - | 5e-29 | 24.06% (478) | polysaccharide biosynthesis protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1495|M.gilvum_PYR-GCK ----MTGSEPEGARPAPEAP-----------STARQTAWNYLVFALSKSS
Mvan_5251|M.vanbaalenii_PYR-1 ------------------MP-----------STGRQTAWNYLIFALSKSS
MSMEG_5968|M.smegmatis_MC2_155 MSSPGGRSDKSEALDPEDLPGAEAPVEPLGRSAAAGTLWLTAAQWLMRLT
* *:. * * * : :
Mflv_1495|M.gilvum_PYR-GCK TLLMTIVVARILSPTEFGIFALAILVVNLFDYVKDLGVGSALVQNRRDWR
Mvan_5251|M.vanbaalenii_PYR-1 TLLMTIVVARILSPTEFGIFALAVLVVNLFDYVKDLGVAAALVQNRREWR
MSMEG_5968|M.smegmatis_MC2_155 GLATIAILTRLLTPEDFGVVAAAMTVTPFLLLLADLGLSTYVLQVKRADQ
* :::*:*:* :**:.* *: *. :: : ***:.: ::* :* :
Mflv_1495|M.gilvum_PYR-GCK VLAPTGLTLSVLFGLFASGVLAATAGVAARALGEPELTELIRVLAVGLAI
Mvan_5251|M.vanbaalenii_PYR-1 ILAPTGLTLSVLFGLGASGVLAATAGVAARALGQPELTGLIRILAVGLAI
MSMEG_5968|M.smegmatis_MC2_155 RLLSTAFWYSLTMAIVLMAAVAAFAPVIASLFDLPKAAPVLRGCSLAAGF
* .*.: *: :.: ..:** * * * :. *: : ::* ::. .:
Mflv_1495|M.gilvum_PYR-GCK SALSTIPAAWLRRSMNFSARLLPEFAGAVAKTGLTIGLAAAGYGVWSLAY
Mvan_5251|M.vanbaalenii_PYR-1 SALSTIPAAWLRRSMNFSARLMPEFAGAATKTGLTIGLAAAGFGVSSLAY
MSMEG_5968|M.smegmatis_MC2_155 IILCSVPAALLRREMRFRALSLQWLASAVIAQVVAVVLAFRGAGAWALVS
*.::*** ***.*.* * : :*.*. ::: ** * *. :*.
Mflv_1495|M.gilvum_PYR-GCK GQLAASVIMTILYWRVARVRPRFAFDGPVCRELVRFGVPVTAVTLLAYAI
Mvan_5251|M.vanbaalenii_PYR-1 GQLAAVVVMTVLYWRVAHIRPSLGFDSRVCRELIRFGLPVTAVTLLAYAI
MSMEG_5968|M.smegmatis_MC2_155 QLVIAEAISCVLVWQAARWRPTLQFSKSEFFDMAKFGNKVIAADVIVAMR
: * .: :* *:.*: ** : *. :: :** * *. ::.
Mflv_1495|M.gilvum_PYR-GCK YNVDYLAVGERLGATELGLYSMAYRIPELLVLSLCVVISEVLFSALSGLQ
Mvan_5251|M.vanbaalenii_PYR-1 YNVDYLAIGERLGATELGLYTMAYRIPELLVLSLCVVVSEVLFSALSGLQ
MSMEG_5968|M.smegmatis_MC2_155 PAAEAAIISTVLGPAALGFLSIAQRLVQVTQDLGGKALVPVSTVVFAKVR
.: :. **.: **: ::* *: :: .: * .:: ::
Mflv_1495|M.gilvum_PYR-GCK DDRDALTGHYLQALTVVAALTIPIGIGLAVAAQPAVGTLYGSAYAESAPI
Mvan_5251|M.vanbaalenii_PYR-1 DDRDALTGHYMQALTVVAALTFPIGVGLAVAAQPLVGTLYGVDYAGSAPI
MSMEG_5968|M.smegmatis_MC2_155 ESPERLQSAYLKALGIAYAAVSPLMTVVVVGGTLVVPLLFGQGWDHSVPV
:. : * . *::** :. * . *: :.*.. * *:* : *.*:
Mflv_1495|M.gilvum_PYR-GCK LSVLAIYATVYSASFHAGDVFKAIGRPGILTAINAGKLAVLVGPIWWAAG
Mvan_5251|M.vanbaalenii_PYR-1 LSVLAIYTALYSASFHAGDVFKAIGRPGILTAINAGKLAVLVVPIWWAAG
MSMEG_5968|M.smegmatis_MC2_155 AQALSLAAILTLGAIIDQRLHYGIGRPGRWLVYAGCIGAVEIAVTALVAH
..*:: : : .:: :. .***** . . ** : .*
Mflv_1495|M.gilvum_PYR-GCK HSALAVAVALLSVEGLHFLVRMTVLCRLTRTSWSSIAGALCRPAAAAVPM
Mvan_5251|M.vanbaalenii_PYR-1 HSALMVAFALLGVEALHFLVRMTVLHRLMGTSWLSIAGALCRPAAAAVPM
MSMEG_5968|M.smegmatis_MC2_155 NGLNWVAIGFVSVAFVATVIRWVLVGRLLDIPPRTLARVFAAASVAVAGS
:. **..::.* : ::* .:: ** . ::* .:. .:.*..
Mflv_1495|M.gilvum_PYR-GCK GLVLVGVGHVIDGLADPLELALMVVVGLCVYVLILRFTAAQLVKAAFEMV
Mvan_5251|M.vanbaalenii_PYR-1 GLVLLGLARVTDGLADPLELVVLAFVGLCVYVLILRFTAAQLVKAAFGMV
MSMEG_5968|M.smegmatis_MC2_155 AGAGLLVLSLTGGWAPAWSLAAVMVAVGVVHIGIVRVVSPDVYRTVRSLV
. . : : : .* * . .*. : .. *:: *:*..:.:: ::. :*
Mflv_1495|M.gilvum_PYR-GCK RARLSRSAATPATTKEDL-
Mvan_5251|M.vanbaalenii_PYR-1 RQRLSRSTAASATTAKEGP
MSMEG_5968|M.smegmatis_MC2_155 HRAPELDESL---------
: . . :