For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARTNSKVRSQYRCSECHHTTAKWVGRCSDCGTWGTVDEVALTAVGGATRRAVAPASPAVPISSIDPGTTR HFHTGISELDRVLGGGLVPGSVTLLAGDPGVGKSTLLLEVAHRWASTGRRALYLSGEESAGQIRLRAERT GCSHDEVYLAAESDLQTALGHIDAVRPSLVVVDSVQTMSTTEADGVTGGVTQVRAVTTALTMTAKTTGVA MLLVGHVTKDGAIAGPRSLEHLVDVVLHFEGDRASALRMVRGVKNRFGAADEVGCFLLHDNGIECVSDPS GLFLDQRPKAVSGTAVTVTLDGKRPMIGEVQALIGASASGSPRRAVSGIDSSRAAMITAVLEKRARLPVG TNDIYLSTVGGMRLTDSSSDLAVALAIASAYTDLALPTTAIAIGEVGLAGDLRRVTGMDKRLAEAARLGF TVAVVPSGAVGAHAGLKVLQADNIGSALQVLRRISQNDASQGRADD*
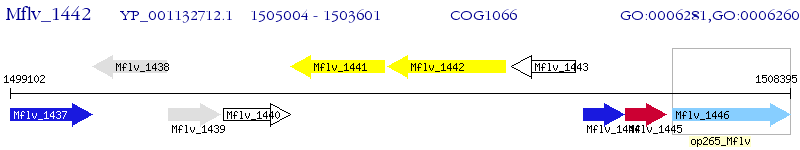
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1442 | - | - | 100% (467) | DNA repair protein RadA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3616 | radA | 0.0 | 72.59% (456) | DNA repair protein RadA |
| M. tuberculosis H37Rv | Rv3585 | radA | 0.0 | 72.59% (456) | DNA repair protein RadA |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0564c | - | 0.0 | 72.22% (450) | DNA repair protein RadA |
| M. marinum M | MMAR_5085 | radA | 0.0 | 70.45% (467) | DNA repair protein RadA |
| M. avium 104 | MAV_0568 | radA | 0.0 | 73.98% (442) | DNA repair protein RadA |
| M. smegmatis MC2 155 | MSMEG_6079 | radA | 0.0 | 80.83% (459) | DNA repair protein RadA |
| M. thermoresistible (build 8) | TH_0587 | radA | 0.0 | 86.63% (389) | DNA REPAIR PROTEIN RADA (DNA REPAIR PROTEIN SMS) |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5342 | - | 0.0 | 93.72% (462) | DNA repair protein RadA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3616|M.bovis_AF2122/97 ----MANARSQYRCSECRHVSAKWVGRCLECGRWGTVDEVAVLSAVGG--
Rv3585|M.tuberculosis_H37Rv ----MANARSQYRCSECRHVSAKWVGRCLECGRWGTVDEVAVLSAVGG--
MMAR_5085|M.marinum_M ----MANARLQYRCADCRYLTAKWVGRCPECSSWGTVEEVSVLSAVGGGS
MAV_0568|M.avium_104 -------------------MTAKWVGRCLECGTWGTVDEVPALSAVAG--
Mflv_1442|M.gilvum_PYR-GCK MARTNSKVRSQYRCSECHHTTAKWVGRCSDCGTWGTVDEVALT-AVG---
Mvan_5342|M.vanbaalenii_PYR-1 MARTNSKTRSQFRCSECHHVTPKWVGRCPDCGTWGTVNEVALT-AVG---
TH_0587|M.thermoresistible__bu --------------------------------------------------
MSMEG_6079|M.smegmatis_MC2_155 MAG--SKIRSQYRCSECQHVAPKWVGRCANCGTWGTVDEVAVL-AGNNKL
MAB_0564c|M.abscessus_ATCC_199 ----MAKPRAQYRCSECQHTTAKWVGRCPDCGTWGSVSETAVLSSIGG--
Mb3616|M.bovis_AF2122/97 --TRRRS--VAPASGAVPISAVDAHRTRPCPTGIDELDRVLGGGIVPGSV
Rv3585|M.tuberculosis_H37Rv --TRRRS--VAPASGAVPISAVDAHRTRPCPTGIDELDRVLGGGIVPGSV
MMAR_5085|M.marinum_M RLGRRAHGPAVPTSQAVPISSVQPNLSRHRPTGIDELDRVLGGGVVPGSV
MAV_0568|M.avium_104 ---RRPR--LTAASQAVPITSIKPDASRHRSTGVDELDRVLGGGVVPGSV
Mflv_1442|M.gilvum_PYR-GCK G-ATRRA--VAPASPAVPISSIDPGTTRHFHTGISELDRVLGGGLVPGSV
Mvan_5342|M.vanbaalenii_PYR-1 GSATRRA--VAPASPAVPISSIDPGSTRHFHTGISELDRVLGGGLVPGSV
TH_0587|M.thermoresistible__bu ---------------------------------VTELDRVLGGGLVPGSV
MSMEG_6079|M.smegmatis_MC2_155 NGAARRS--VAPTSPAVPITSIDPGVTRHYPTGVSELDRVLGGGLVAGSV
MAB_0564c|M.abscessus_ATCC_199 -LGAVRA--VAPASAAVPITSIATDTTRHQPTGVSELDRVLGGGLIPGSV
: *********::.***
Mb3616|M.bovis_AF2122/97 TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYVSGEESAGQIRLRADRIGC
Rv3585|M.tuberculosis_H37Rv TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYVSGEESAGQIRLRADRIGC
MMAR_5085|M.marinum_M TLLAGEPGVGKSTLLLEVAHRWAQSGQRVLYISGEESAGQIRLRADRIGC
MAV_0568|M.avium_104 TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYISGEESAGQIRLRADRIGC
Mflv_1442|M.gilvum_PYR-GCK TLLAGDPGVGKSTLLLEVAHRWASTGRRALYLSGEESAGQIRLRAERTGC
Mvan_5342|M.vanbaalenii_PYR-1 TLLAGDPGVGKSTLLLEVAHRWASTGRRALYLSGEESAGQIRLRAERTGC
TH_0587|M.thermoresistible__bu TLLAGDPGVGKSTLLLEVAHRWARAGRRALYLSGEESAGQIRLRAERTGC
MSMEG_6079|M.smegmatis_MC2_155 TLLAGDPGVGKSTLLLEVANRWAHSGKRALYLSGEESAGQIRLRAERTGC
MAB_0564c|M.abscessus_ATCC_199 NLLAGEPGVGKSTLLLKVVHQWAASGGRALYISGEESAAQVRLRAERTGC
.****:**********:*.::** :* *.**:******.*:****:* **
Mb3616|M.bovis_AF2122/97 GT-------------EVEEIYLAAQSDVHTVLDQIETVQPALVIVDSVQT
Rv3585|M.tuberculosis_H37Rv GT-------------EVEEIYLAAQSDVHTVLDQIETVQPALVIVDSVQT
MMAR_5085|M.marinum_M GGPAAPDISEVSAIDNSAEIYLAAESDLHAVLDHIDTVGSALVIVDSVQT
MAV_0568|M.avium_104 GG---------------DQVYLAAESDLHTVLEHVATVKPALVIVDSVQT
Mflv_1442|M.gilvum_PYR-GCK SH---------------DEVYLAAESDLQTALGHIDAVRPSLVVVDSVQT
Mvan_5342|M.vanbaalenii_PYR-1 SH---------------DEVYLAAESDLQTALGHIDAVQPTLVVVDSVQT
TH_0587|M.thermoresistible__bu TH---------------DEVFLAAESDLQTALGHIDAVQPSLVVVDSVQT
MSMEG_6079|M.smegmatis_MC2_155 TH---------------DQVYLAAESDLQIALGHIDEVKPSLVVVDSVQT
MAB_0564c|M.abscessus_ATCC_199 VH---------------DEVYLASESDLHSVLGHIEAVKPTLAIIDSVQT
:::**::**:: .* :: * .:*.::*****
Mb3616|M.bovis_AF2122/97 MSTSEADGVTGGVTQVRAVTAALTAAAKA----NEVALILVGHVTKDGAI
Rv3585|M.tuberculosis_H37Rv MSTSEADGVTGGVTQVRAVTAALTAAAKA----NEVALILVGHVTKDGAI
MMAR_5085|M.marinum_M MSTSEVDGVTGGVTQVRAVTAALTAAAKS----NGFALILVGHVTKDGAI
MAV_0568|M.avium_104 MSTTEADGVAGGVTQVRAVTAALTAAAKA----NGVALILVGHVTKDGAI
Mflv_1442|M.gilvum_PYR-GCK MSTTEADGVTGGVTQVRAVTTALTMTAKT----TGVAMLLVGHVTKDGAI
Mvan_5342|M.vanbaalenii_PYR-1 MSTTEADGVTGGVTQVRAVTTALTMTAKT----TGVAMLLVGHVTKDGAI
TH_0587|M.thermoresistible__bu MSTTEADGVTGGVTQVRAVTTALTSYAKASA-APGVAMLLVGHVTKDGAI
MSMEG_6079|M.smegmatis_MC2_155 MSTTEADGVTGGVTQVRAVTTSLTAYAKAAVGDPAVAMILVGHVTKDGAI
MAB_0564c|M.abscessus_ATCC_199 MSTTDADGVTGGVTQVRAVTTALTMAAKS----SGVAMVLVGHVTKDGAI
***::.***:**********::** **: .*::***********
Mb3616|M.bovis_AF2122/97 AGPRSLEHLVDVVLHFEGDRNGALRMVRGVKNRFGAADEVGCFLLHDNGI
Rv3585|M.tuberculosis_H37Rv AGPRSLEHLVDVVLHFEGDRNGALRMVRGVKNRFGAADEVGCFLLHDNGI
MMAR_5085|M.marinum_M AGPRSLEHLVDVVLHFEGDRNGGLRMVRGVKNRFGAADEVGCFLLHDNGI
MAV_0568|M.avium_104 AGPRSLEHLVDVVLHFEGDRNGSLRMVRGIKNRFGAADEVGCFLLHDNGI
Mflv_1442|M.gilvum_PYR-GCK AGPRSLEHLVDVVLHFEGDRASALRMVRGVKNRFGAADEVGCFLLHDNGI
Mvan_5342|M.vanbaalenii_PYR-1 AGPRSLEHLVDVVLHFEGDRASALRMVRGIKNRFGAADEVGCFLLHDNGI
TH_0587|M.thermoresistible__bu AGPRSLEHLVDVVLHFEGDRTTALRMVRGVKNRFGAADEVGCFLLHDSGI
MSMEG_6079|M.smegmatis_MC2_155 AGPRSLEHLVDVVLHFEGDRASSLRMVRGVKNRFGAADEVGCFQLHDNGI
MAB_0564c|M.abscessus_ATCC_199 AGPRSLEHLVDVVLHFEGDKHSALRMVRGIKNRFGAADEVGCFQLRDKGI
*******************: .******:************* *:*.**
Mb3616|M.bovis_AF2122/97 DGIVDPSNLFLDQRPTPVAGTAITVTLDGKRPLVGEVQALLATPCGGSPR
Rv3585|M.tuberculosis_H37Rv DGIVDPSNLFLDQRPTPVAGTAITVTLDGKRPLVGEVQALLATPCGGSPR
MMAR_5085|M.marinum_M EGVADPSNLFLDQRPTPVPGTAITVTLDGKRPLIGEVQALLATPNGGAPR
MAV_0568|M.avium_104 AGVTDPSNLFLDQRSAPVPGTAITVTLDGKRPLIGEVQALLASPSGGSPR
Mflv_1442|M.gilvum_PYR-GCK ECVSDPSGLFLDQRPKAVSGTAVTVTLDGKRPMIGEVQALIGASASGSPR
Mvan_5342|M.vanbaalenii_PYR-1 ECVSDPSGLFLDQRPKPVSGTAVTVTLDGKRPMIGEVQALIGSPASGSPR
TH_0587|M.thermoresistible__bu ECVVDPSGLFLDQRPAPVSGTAVTVTLDGKRPLIGEVQALIGAPATGTPR
MSMEG_6079|M.smegmatis_MC2_155 ECVSDPSGLFLDQRPLAVPGTAVTVTLDGKRPMIGEVQALVSPPA-GPPR
MAB_0564c|M.abscessus_ATCC_199 ECVSDPSGLFLEHREKAAAGTAVTVTLDGKRPLLGEVQSLVTTSTNPSPR
: ***.***::* ...***:*********::****:*: .. .**
Mb3616|M.bovis_AF2122/97 RAVSGIHQARAAMIAAVLEKHARLAIAVNDIYLSTVGGMRLTEPSADLAV
Rv3585|M.tuberculosis_H37Rv RAVSGIHQARAAMIAAVLEKHARLAIAVNDIYLSTVGGMRLTEPSADLAV
MMAR_5085|M.marinum_M RAVSGIDHARAAMISAVLEKHARLSLAVNDMYLSTVGGMRLTDPSSDLAV
MAV_0568|M.avium_104 RAVSGIDHSRAAMITAVLEKHGKLPVAVNDIYLSTVGGMRLTDPSSDLAV
Mflv_1442|M.gilvum_PYR-GCK RAVSGIDSSRAAMITAVLEKRARLPVGTNDIYLSTVGGMRLTDSSSDLAV
Mvan_5342|M.vanbaalenii_PYR-1 RAVSGIDSSRAAMITAVLEKRAKLPVGMNDIYLSTVGGMRLTDSSSDLAV
TH_0587|M.thermoresistible__bu RAVSGIDSARAAMITAVLEKRARLPVGSNDIYLSTVGGMRLTDSSSDLAV
MSMEG_6079|M.smegmatis_MC2_155 RAVSGIDSARAAMIGAVLQTRCRMPINSNDLYLSTVGGMRLTDPSADLAV
MAB_0564c|M.abscessus_ATCC_199 RAVSGLDHARSAMVTAVLERHGRLPLGNNDIYMATVGGMRMTDPSADLAI
*****:. :*:**: ***: : ::.: **:*::******:*:.*:***:
Mb3616|M.bovis_AF2122/97 AIALASAYANLPLPTTAVMIGEVGLAGDIRRVNGMARRLSEAARQGFTIA
Rv3585|M.tuberculosis_H37Rv AIALASAYANLPLPTTAVMIGEVGLAGDIRRVNGMARRLSEAARQGFTIA
MMAR_5085|M.marinum_M AIALASAYANLPLPTTAVMIGEVGLAGDLRRVNGMDRRLAEAARQGFNIA
MAV_0568|M.avium_104 AIALASALSELPLPTTAVMIGEVGLAGDLRRVSGMARRLSEAARQGFSIA
Mflv_1442|M.gilvum_PYR-GCK ALAIASAYTDLALPTTAIAIGEVGLAGDLRRVTGMDKRLAEAARLGFTVA
Mvan_5342|M.vanbaalenii_PYR-1 ALSIASAFTDLALPTTAIAIGEVGLAGDLRRVTGMDRRLAEAARLGFTYA
TH_0587|M.thermoresistible__bu ALAIASAYTDLPLPTTAVAIGEVGLAGDLRRVTGMDRRLAEAARLGFGTA
MSMEG_6079|M.smegmatis_MC2_155 ALAIASAYFDIAMPMKAIAIGEVGLAGDLRRVTGMDRRLSEAARLGFTTA
MAB_0564c|M.abscessus_ATCC_199 LMAVASAYTDIALPANMVFIGEVGLAGDIRRTAAVGKRISEAARLGYTHA
:::*** ::.:* . : *********:**. .: :*::**** *: *
Mb3616|M.bovis_AF2122/97 LVPPSD----DPVPPGMHALRASTIVAALQYMVDIADHRGTTLATPPSHS
Rv3585|M.tuberculosis_H37Rv LVPPSD----DPVPPGMHALRASTIVAALQYMVDIADHRGTTLATPPSHS
MMAR_5085|M.marinum_M LVPPG----CEAVPAGLRTLCAPTIVAALEHMIDIADHR-SVPSAKVHRL
MAV_0568|M.avium_104 LIPDGDDPRREIVPSGMRALRAPTIGAALQHMVDIAEPR-KGWTHRALGP
Mflv_1442|M.gilvum_PYR-GCK VVPSG----AVGAHAGLKVLQADNIGSALQVLRRISQNDASQGRADD---
Mvan_5342|M.vanbaalenii_PYR-1 VVPSG----VTSAPAGLQVLPADNITSALQVLRRISQNGAN---------
TH_0587|M.thermoresistible__bu VIPPG----VTSVPRGLRAITADNITAALRVLRDIADNGGRQ--------
MSMEG_6079|M.smegmatis_MC2_155 VVPPG----VTSAPAGLKVVAADNIRAAVQTMREIAIAGAQ---------
MAB_0564c|M.abscessus_ATCC_199 VIPPEG---DDLHPAGMHLLRCPTVGRALEVLSEARKIEPEKRPNLRLAQ
::* *:: : . .: *:. :
Mb3616|M.bovis_AF2122/97 GTGHVPLGRGT
Rv3585|M.tuberculosis_H37Rv GTGHVPLGRGT
MMAR_5085|M.marinum_M DTSH-------
MAV_0568|M.avium_104 QNDTL------
Mflv_1442|M.gilvum_PYR-GCK -----------
Mvan_5342|M.vanbaalenii_PYR-1 -----------
TH_0587|M.thermoresistible__bu -----------
MSMEG_6079|M.smegmatis_MC2_155 -----------
MAB_0564c|M.abscessus_ATCC_199 QFTD-------