For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAGSKIRSQYRCSECQHVAPKWVGRCANCGTWGTVDEVAVLAGNNKLNGAARRSVAPTSPAVPITSIDPG VTRHYPTGVSELDRVLGGGLVAGSVTLLAGDPGVGKSTLLLEVANRWAHSGKRALYLSGEESAGQIRLRA ERTGCTHDQVYLAAESDLQIALGHIDEVKPSLVVVDSVQTMSTTEADGVTGGVTQVRAVTTSLTAYAKAA VGDPAVAMILVGHVTKDGAIAGPRSLEHLVDVVLHFEGDRASSLRMVRGVKNRFGAADEVGCFQLHDNGI ECVSDPSGLFLDQRPLAVPGTAVTVTLDGKRPMIGEVQALVSPPAGPPRRAVSGIDSARAAMIGAVLQTR CRMPINSNDLYLSTVGGMRLTDPSADLAVALAIASAYFDIAMPMKAIAIGEVGLAGDLRRVTGMDRRLSE AARLGFTTAVVPPGVTSAPAGLKVVAADNIRAAVQTMREIAIAGAQ
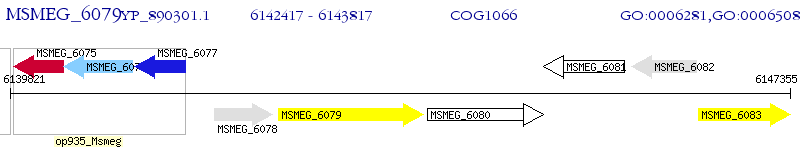
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6079 | radA | - | 100% (466) | DNA repair protein RadA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3616 | radA | 1e-178 | 70.74% (458) | DNA repair protein RadA |
| M. gilvum PYR-GCK | Mflv_1442 | - | 0.0 | 80.83% (459) | DNA repair protein RadA |
| M. tuberculosis H37Rv | Rv3585 | radA | 1e-178 | 70.74% (458) | DNA repair protein RadA |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0564c | - | 1e-178 | 68.12% (458) | DNA repair protein RadA |
| M. marinum M | MMAR_5085 | radA | 1e-176 | 67.30% (474) | DNA repair protein RadA |
| M. avium 104 | MAV_0568 | radA | 1e-170 | 70.38% (449) | DNA repair protein RadA |
| M. thermoresistible (build 8) | TH_0587 | radA | 1e-178 | 82.01% (389) | DNA REPAIR PROTEIN RADA (DNA REPAIR PROTEIN SMS) |
| M. ulcerans Agy99 | MUL_0075 | dnaB | 2e-05 | 42.86% (56) | replicative DNA helicase DnaB |
| M. vanbaalenii PYR-1 | Mvan_5342 | - | 0.0 | 80.13% (463) | DNA repair protein RadA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1442|M.gilvum_PYR-GCK MARTNSKVRSQYRCSECHHTTAKWVGRCSDCGTWGTVDEVALT-AVG---
Mvan_5342|M.vanbaalenii_PYR-1 MARTNSKTRSQFRCSECHHVTPKWVGRCPDCGTWGTVNEVALT-AVG---
TH_0587|M.thermoresistible__bu --------------------------------------------------
MSMEG_6079|M.smegmatis_MC2_155 MAG--SKIRSQYRCSECQHVAPKWVGRCANCGTWGTVDEVAVL-AGNNKL
MMAR_5085|M.marinum_M ----MANARLQYRCADCRYLTAKWVGRCPECSSWGTVEEVSVLSAVGGGS
MAV_0568|M.avium_104 -------------------MTAKWVGRCLECGTWGTVDEVPALSAVAG--
Mb3616|M.bovis_AF2122/97 ----MANARSQYRCSECRHVSAKWVGRCLECGRWGTVDEVAVLSAVGG--
Rv3585|M.tuberculosis_H37Rv ----MANARSQYRCSECRHVSAKWVGRCLECGRWGTVDEVAVLSAVGG--
MAB_0564c|M.abscessus_ATCC_199 ----MAKPRAQYRCSECQHTTAKWVGRCPDCGTWGSVSETAVLSSIGG--
MUL_0075|M.ulcerans_Agy99 ---------MAVVDDLAHSDNMDAPPPSEDFGRQPPQDLAAEQSVLGG--
Mflv_1442|M.gilvum_PYR-GCK G-ATRRA--VAPASPAVPISSIDPGTTRHFHTGISELDRVLGGGLVPGSV
Mvan_5342|M.vanbaalenii_PYR-1 GSATRRA--VAPASPAVPISSIDPGSTRHFHTGISELDRVLGGGLVPGSV
TH_0587|M.thermoresistible__bu ---------------------------------VTELDRVLGGGLVPGSV
MSMEG_6079|M.smegmatis_MC2_155 NGAARRS--VAPTSPAVPITSIDPGVTRHYPTGVSELDRVLGGGLVAGSV
MMAR_5085|M.marinum_M RLGRRAHGPAVPTSQAVPISSVQPNLSRHRPTGIDELDRVLGGGVVPGSV
MAV_0568|M.avium_104 ---RRPR--LTAASQAVPITSIKPDASRHRSTGVDELDRVLGGGVVPGSV
Mb3616|M.bovis_AF2122/97 --TRRRS--VAPASGAVPISAVDAHRTRPCPTGIDELDRVLGGGIVPGSV
Rv3585|M.tuberculosis_H37Rv --TRRRS--VAPASGAVPISAVDAHRTRPCPTGIDELDRVLGGGIVPGSV
MAB_0564c|M.abscessus_ATCC_199 -LGAVRA--VAPASAAVPITSIATDTTRHQPTGVSELDRVLGGGLIPGSV
MUL_0075|M.ulcerans_Agy99 --------MLLSKDAVADVLERLRPGDFYRPAHQNIYDAILDLYGRGEPA
* :*. ..
Mflv_1442|M.gilvum_PYR-GCK TLLAGDPGVGKSTLLLEVAHRWASTGRRALYLSGEESAGQIRLRAERTGC
Mvan_5342|M.vanbaalenii_PYR-1 TLLAGDPGVGKSTLLLEVAHRWASTGRRALYLSGEESAGQIRLRAERTGC
TH_0587|M.thermoresistible__bu TLLAGDPGVGKSTLLLEVAHRWARAGRRALYLSGEESAGQIRLRAERTGC
MSMEG_6079|M.smegmatis_MC2_155 TLLAGDPGVGKSTLLLEVANRWAHSGKRALYLSGEESAGQIRLRAERTGC
MMAR_5085|M.marinum_M TLLAGEPGVGKSTLLLEVAHRWAQSGQRVLYISGEESAGQIRLRADRIGC
MAV_0568|M.avium_104 TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYISGEESAGQIRLRADRIGC
Mb3616|M.bovis_AF2122/97 TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYVSGEESAGQIRLRADRIGC
Rv3585|M.tuberculosis_H37Rv TLLAGDPGVGKSTLLLEVAHRWAQSGRRALYVSGEESAGQIRLRADRIGC
MAB_0564c|M.abscessus_ATCC_199 NLLAGEPGVGKSTLLLKVVHQWAASGGRALYISGEESAAQVRLRAERTGC
MUL_0075|M.ulcerans_Agy99 DAVTVAAELDRRGLLRRIGGAPYLHTLISTVPTAANAGYYAGIVGEKALL
:: . :.: ** .: :. ::. : .::
Mflv_1442|M.gilvum_PYR-GCK SH---------------DEVYLAAESDLQTALGHIDAVRPSLVVVDSVQT
Mvan_5342|M.vanbaalenii_PYR-1 SH---------------DEVYLAAESDLQTALGHIDAVQPTLVVVDSVQT
TH_0587|M.thermoresistible__bu TH---------------DEVFLAAESDLQTALGHIDAVQPSLVVVDSVQT
MSMEG_6079|M.smegmatis_MC2_155 TH---------------DQVYLAAESDLQIALGHIDEVKPSLVVVDSVQT
MMAR_5085|M.marinum_M GGPAAPDISEVSAIDNSAEIYLAAESDLHAVLDHIDTVGSALVIVDSVQT
MAV_0568|M.avium_104 GG---------------DQVYLAAESDLHTVLEHVATVKPALVIVDSVQT
Mb3616|M.bovis_AF2122/97 GT-------------EVEEIYLAAQSDVHTVLDQIETVQPALVIVDSVQT
Rv3585|M.tuberculosis_H37Rv GT-------------EVEEIYLAAQSDVHTVLDQIETVQPALVIVDSVQT
MAB_0564c|M.abscessus_ATCC_199 VH---------------DEVYLASESDLHSVLGHIEAVKPTLAIIDSVQT
MUL_0075|M.ulcerans_Agy99 RR-----------------LVEAGTRVVQYGYAGAEGADVAEVVDRAQAE
: *. :: . : .: :
Mflv_1442|M.gilvum_PYR-GCK MSTTEADGVTGGVTQVRAVTTALTMTAKT----TGVAM-LLVGHVTKDGA
Mvan_5342|M.vanbaalenii_PYR-1 MSTTEADGVTGGVTQVRAVTTALTMTAKT----TGVAM-LLVGHVTKDGA
TH_0587|M.thermoresistible__bu MSTTEADGVTGGVTQVRAVTTALTSYAKASA-APGVAM-LLVGHVTKDGA
MSMEG_6079|M.smegmatis_MC2_155 MSTTEADGVTGGVTQVRAVTTSLTAYAKAAVGDPAVAM-ILVGHVTKDGA
MMAR_5085|M.marinum_M MSTSEVDGVTGGVTQVRAVTAALTAAAKS----NGFAL-ILVGHVTKDGA
MAV_0568|M.avium_104 MSTTEADGVAGGVTQVRAVTAALTAAAKA----NGVAL-ILVGHVTKDGA
Mb3616|M.bovis_AF2122/97 MSTSEADGVTGGVTQVRAVTAALTAAAKA----NEVAL-ILVGHVTKDGA
Rv3585|M.tuberculosis_H37Rv MSTSEADGVTGGVTQVRAVTAALTAAAKA----NEVAL-ILVGHVTKDGA
MAB_0564c|M.abscessus_ATCC_199 MSTTDADGVTGGVTQVRAVTTALTMAAKS----SGVAM-VLVGHVTKDGA
MUL_0075|M.ulcerans_Agy99 IYDVADRRLSEDFVPLEDLLQPTMDEIDAIASNGGISRGVPTGFTELDEV
: :: ... :. : . .: .: : .*.. * .
Mflv_1442|M.gilvum_PYR-GCK IAGPRSLEHLVDVVLHFEGDRASALRMVRGVKNRFGAADEVGCFLLHDNG
Mvan_5342|M.vanbaalenii_PYR-1 IAGPRSLEHLVDVVLHFEGDRASALRMVRGIKNRFGAADEVGCFLLHDNG
TH_0587|M.thermoresistible__bu IAGPRSLEHLVDVVLHFEGDRTTALRMVRGVKNRFGAADEVGCFLLHDSG
MSMEG_6079|M.smegmatis_MC2_155 IAGPRSLEHLVDVVLHFEGDRASSLRMVRGVKNRFGAADEVGCFQLHDNG
MMAR_5085|M.marinum_M IAGPRSLEHLVDVVLHFEGDRNGGLRMVRGVKNRFGAADEVGCFLLHDNG
MAV_0568|M.avium_104 IAGPRSLEHLVDVVLHFEGDRNGSLRMVRGIKNRFGAADEVGCFLLHDNG
Mb3616|M.bovis_AF2122/97 IAGPRSLEHLVDVVLHFEGDRNGALRMVRGVKNRFGAADEVGCFLLHDNG
Rv3585|M.tuberculosis_H37Rv IAGPRSLEHLVDVVLHFEGDRNGALRMVRGVKNRFGAADEVGCFLLHDNG
MAB_0564c|M.abscessus_ATCC_199 IAGPRSLEHLVDVVLHFEGDKHSALRMVRGIKNRFGAADEVGCFQLRDKG
MUL_0075|M.ulcerans_Agy99 TNGLHPGQMIIVAARPGVGKSTLGLDFMRSCSIKHQMASVIFSLEMSKTE
* :. : :: .. *. .* ::*. . :. *. : .: : ..
Mflv_1442|M.gilvum_PYR-GCK IEC---VSDPSGLFLDQRPKAVSGTAVTVTLDGKRPMIGEVQALIGASAS
Mvan_5342|M.vanbaalenii_PYR-1 IEC---VSDPSGLFLDQRPKPVSGTAVTVTLDGKRPMIGEVQALIGSPAS
TH_0587|M.thermoresistible__bu IEC---VVDPSGLFLDQRPAPVSGTAVTVTLDGKRPLIGEVQALIGAPAT
MSMEG_6079|M.smegmatis_MC2_155 IEC---VSDPSGLFLDQRPLAVPGTAVTVTLDGKRPMIGEVQALVSPPA-
MMAR_5085|M.marinum_M IEG---VADPSNLFLDQRPTPVPGTAITVTLDGKRPLIGEVQALLATPNG
MAV_0568|M.avium_104 IAG---VTDPSNLFLDQRSAPVPGTAITVTLDGKRPLIGEVQALLASPSG
Mb3616|M.bovis_AF2122/97 IDG---IVDPSNLFLDQRPTPVAGTAITVTLDGKRPLVGEVQALLATPCG
Rv3585|M.tuberculosis_H37Rv IDG---IVDPSNLFLDQRPTPVAGTAITVTLDGKRPLVGEVQALLATPCG
MAB_0564c|M.abscessus_ATCC_199 IEC---VSDPSGLFLEHREKAAAGTAVTVTLDGKRPLLGEVQSLVTTSTN
MUL_0075|M.ulcerans_Agy99 IVMRLLSAEAKIKLADMRSGRMTDDDWTRLARRMSEISEAPLYIDDSPNL
* :.. : : * .. * : : ..
Mflv_1442|M.gilvum_PYR-GCK GSPRRAVSGIDSSRAAMITAVLEKRARLPVGTNDIYLSTVGGMRLTDSSS
Mvan_5342|M.vanbaalenii_PYR-1 GSPRRAVSGIDSSRAAMITAVLEKRAKLPVGMNDIYLSTVGGMRLTDSSS
TH_0587|M.thermoresistible__bu GTPRRAVSGIDSARAAMITAVLEKRARLPVGSNDIYLSTVGGMRLTDSSS
MSMEG_6079|M.smegmatis_MC2_155 GPPRRAVSGIDSARAAMIGAVLQTRCRMPINSNDLYLSTVGGMRLTDPSA
MMAR_5085|M.marinum_M GAPRRAVSGIDHARAAMISAVLEKHARLSLAVNDMYLSTVGGMRLTDPSS
MAV_0568|M.avium_104 GSPRRAVSGIDHSRAAMITAVLEKHGKLPVAVNDIYLSTVGGMRLTDPSS
Mb3616|M.bovis_AF2122/97 GSPRRAVSGIHQARAAMIAAVLEKHARLAIAVNDIYLSTVGGMRLTEPSA
Rv3585|M.tuberculosis_H37Rv GSPRRAVSGIHQARAAMIAAVLEKHARLAIAVNDIYLSTVGGMRLTEPSA
MAB_0564c|M.abscessus_ATCC_199 PSPRRAVSGLDHARSAMVTAVLERHGRLPLGNNDIYMATVGGMRMTDPSA
MUL_0075|M.ulcerans_Agy99 TTMEIRAKARRLRQKANLKLVVVDYLQLMISGKKHESRQVEVSEFSRHLK
. . ... : * : *: :: : :. * .::
Mflv_1442|M.gilvum_PYR-GCK DLAVALAIASAYTDLALPTTAIAIGEVGLAGDLRRVTGMDKRLAEAARLG
Mvan_5342|M.vanbaalenii_PYR-1 DLAVALSIASAFTDLALPTTAIAIGEVGLAGDLRRVTGMDRRLAEAARLG
TH_0587|M.thermoresistible__bu DLAVALAIASAYTDLPLPTTAVAIGEVGLAGDLRRVTGMDRRLAEAARLG
MSMEG_6079|M.smegmatis_MC2_155 DLAVALAIASAYFDIAMPMKAIAIGEVGLAGDLRRVTGMDRRLSEAARLG
MMAR_5085|M.marinum_M DLAVAIALASAYANLPLPTTAVMIGEVGLAGDLRRVNGMDRRLAEAARQG
MAV_0568|M.avium_104 DLAVAIALASALSELPLPTTAVMIGEVGLAGDLRRVSGMARRLSEAARQG
Mb3616|M.bovis_AF2122/97 DLAVAIALASAYANLPLPTTAVMIGEVGLAGDIRRVNGMARRLSEAARQG
Rv3585|M.tuberculosis_H37Rv DLAVAIALASAYANLPLPTTAVMIGEVGLAGDIRRVNGMARRLSEAARQG
MAB_0564c|M.abscessus_ATCC_199 DLAILMAVASAYTDIALPANMVFIGEVGLAGDIRRTAAVGKRISEAARLG
MUL_0075|M.ulcerans_Agy99 LLAKEIEVPVVAISQLNRGPEQRTDKKPMLSDLRESGSLEQDADMVILLN
** : :. . . .: : .*:*. .: : . .
Mflv_1442|M.gilvum_PYR-GCK FTVAVVPSG----AVGAHAGLKVLQADNIGSALQVLRRISQNDASQGRAD
Mvan_5342|M.vanbaalenii_PYR-1 FTYAVVPSG----VTSAPAGLQVLPADNITSALQVLRRISQNGAN-----
TH_0587|M.thermoresistible__bu FGTAVIPPG----VTSVPRGLRAITADNITAALRVLRDIADNGGRQ----
MSMEG_6079|M.smegmatis_MC2_155 FTTAVVPPG----VTSAPAGLKVVAADNIRAAVQTMREIAIAGAQ-----
MMAR_5085|M.marinum_M FNIALVPPG----CEAVPAGLRTLCAPTIVAALEHMIDIADHR-SVPSAK
MAV_0568|M.avium_104 FSIALIPDGDDPRREIVPSGMRALRAPTIGAALQHMVDIAEPR-KGWTHR
Mb3616|M.bovis_AF2122/97 FTIALVPPS----DDPVPPGMHALRASTIVAALQYMVDIADHRGTTLATP
Rv3585|M.tuberculosis_H37Rv FTIALVPPS----DDPVPPGMHALRASTIVAALQYMVDIADHRGTTLATP
MAB_0564c|M.abscessus_ATCC_199 YTHAVIPPEG---DDLHPAGMHLLRCPTVGRALEVLSEARKIEPEKRPNL
MUL_0075|M.ulcerans_Agy99 RPDAFERDD------PRGGEADFILAKHRNGPTKTVTVAHQLHLSRFANM
*. : . . . :
Mflv_1442|M.gilvum_PYR-GCK D--------------
Mvan_5342|M.vanbaalenii_PYR-1 ---------------
TH_0587|M.thermoresistible__bu ---------------
MSMEG_6079|M.smegmatis_MC2_155 ---------------
MMAR_5085|M.marinum_M VHRLDTSH-------
MAV_0568|M.avium_104 ALGPQNDTL------
Mb3616|M.bovis_AF2122/97 PSHSGTGHVPLGRGT
Rv3585|M.tuberculosis_H37Rv PSHSGTGHVPLGRGT
MAB_0564c|M.abscessus_ATCC_199 RLAQQFTD-------
MUL_0075|M.ulcerans_Agy99 AR-------------