For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MITFENVTKRYPDGTVAVDDLTLEVPEGTLTVFVGPSGCGKTTSMRMINRMIEPTSGTLTVDGKDVTKVD PVKLRLGIGYVIQSAGLMPHLRVIDNVATVPVLQGESRRSARKSALGVMERVGLDPKLADRYPAQLSGGQ QQRVGVARALAADPPILLMDEPFSAVDPVVRDDLQAEIVRLQSELRKTIVFVTHDIDEAIKLGDRVAVFG RGGVLEQYDAPARLLSNPANDSVAGFVGADRGYRGLQFFHATGLPLHDIRHVGEPEIDALTLGSGDWVLV TRPDGTPYAWIDAEGVDLHRKGASLYDSTIAGGSLFRPDGTLRLALDAALSSPSGLGVAVDADGQVVGGV KADDVLAALDTQRRARHEG
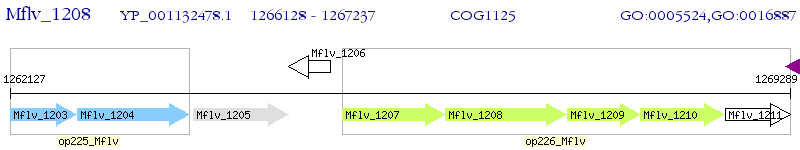
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1208 | - | - | 100% (369) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_3109 | - | 1e-78 | 44.39% (374) | glycine betaine/L-proline ABC transporter, ATPase subunit |
| M. gilvum PYR-GCK | Mflv_2681 | - | 8e-48 | 41.11% (270) | sulfate ABC transporter, ATPase subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3784c | proV | 1e-135 | 67.85% (367) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. tuberculosis H37Rv | Rv3758c | proV | 1e-135 | 67.85% (367) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | MLBr_01089 | - | 3e-44 | 41.60% (238) | putative ABC-transport protein, ATP-binding component |
| M. abscessus ATCC 19977 | MAB_0262 | - | 1e-161 | 77.75% (364) | amino acid ABC transporter ATP-binding protein |
| M. marinum M | MMAR_5301 | proV | 1e-135 | 67.49% (363) | osmoprotectant transport ATP-binding protein ABC transporter |
| M. avium 104 | MAV_0318 | - | 1e-137 | 68.58% (366) | amino acid ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_6333 | - | 0.0 | 86.81% (364) | amino acid ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4520 | - | 1e-81 | 46.34% (369) | PUTATIVE osmoprotectant (glycine |
| M. ulcerans Agy99 | MUL_0033 | proV_1 | 3e-74 | 44.20% (371) | osmoprotectant (glycine |
| M. vanbaalenii PYR-1 | Mvan_5600 | - | 0.0 | 90.16% (366) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3784c|M.bovis_AF2122/97 ---------------------MICFDDVSKVY-AHGATAVDRLTLEVPNG
Rv3758c|M.tuberculosis_H37Rv ---------------------MICFDDVSKVY-AHGATAVDRLTLEVPNG
MMAR_5301|M.marinum_M ---------------------MIRFDNVDKVY-ADGTTAVDQLSLEVADG
MAV_0318|M.avium_104 ---------------------MIVFDNVSKVF-ADGTAAVDRLSLTVPTG
Mflv_1208|M.gilvum_PYR-GCK ---------------------MITFENVTKRY-PDGTVAVDDLTLEVPEG
Mvan_5600|M.vanbaalenii_PYR-1 ---------------------MISFEHVTKRY-PDGTVAVDDLTLEVPEG
MSMEG_6333|M.smegmatis_MC2_155 ---------------------MITFTDVTKQY-PDGTVAVDNLNLEVPQG
MAB_0262|M.abscessus_ATCC_1997 ---------------------MITFENVTKQY-PDGTTAVDDLNMHVDKG
TH_4520|M.thermoresistible__bu ------VSEP-TSDLQSGGGARILLDGVTKRYDPKSPPAVDNITMEIPAG
MUL_0033|M.ulcerans_Agy99 MPEAASADQPTTSDAPQSSGLRILLDGVTKRYGVRSTPAVDNVTLEIGAG
MLBr_01089|M.leprae_Br4923 -------------------MAEIVLEHVNKNY-PNGATAVHDLSITVADG
* : * * : .. **. :.: : *
Mb3784c|M.bovis_AF2122/97 MLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLRLGI
Rv3758c|M.tuberculosis_H37Rv MLTVFVGPSGCGKTTALRMINRMVDPTSGTITVDGTDVSTVNAVKLRLGI
MMAR_5301|M.marinum_M MLTVFVGPSGCGKTTALRMINRMVEPSSGTVTVNGTDVSTVNPVKLRLGI
MAV_0318|M.avium_104 KLTVFVGSSGSGKTTALRMINRMIEPTSGTISIDGVDVSTLDPVKLRLGI
Mflv_1208|M.gilvum_PYR-GCK TLTVFVGPSGCGKTTSMRMINRMIEPTSGTLTVDGKDVTKVDPVKLRLGI
Mvan_5600|M.vanbaalenii_PYR-1 TLAVFVGPSGCGKTTSMRMINRMIEPTSGTLTVDGRDVTKVDAVKLRLGI
MSMEG_6333|M.smegmatis_MC2_155 TLAAFVGPSGCGKTTSMRMVNRMIDPTSGTLTVNGEDVTQVDPVKLRLGI
MAB_0262|M.abscessus_ATCC_1997 SFTVFVGPSGCGKTTSMRMINRMTDLTSGLLTVDGADVRTVDPVKLRLGI
TH_4520|M.thermoresistible__bu EIVMLVGPSGCGKTTTMKMINRLIEPTSGRIFIGDDDVTERDADELRRHI
MUL_0033|M.ulcerans_Agy99 EIVMLVGPSGCGKTTTMKVINRLIEPTSGRIFIGDDDVTRRNPDELRRHI
MLBr_01089|M.leprae_Br4923 EFLILIGPSGCGKTTTLNMIAGLEDISSGELRIDGDRVNEKAPK--DRDI
: ::*.**.****::.:: : : :** : :.. * . *
Mb3784c|M.bovis_AF2122/97 GYVIQNAGLMPHQRVIDNVATVPVLKGQPRRAARKAGYEVLERVGLDP-K
Rv3758c|M.tuberculosis_H37Rv GYVIQNAGLMPHQRVIDNVATVPVLKGQPRRAARKAGYEVLERVGLDP-K
MMAR_5301|M.marinum_M GYVIQNAGLMPHQRVIDNVATVPVLKGQSRRAARKAAYEVLERVGLNP-K
MAV_0318|M.avium_104 GYVIQHAGLMPHQRVIDNVATVPVLRGQRRRAARTAAYQVLERVGLDV-K
Mflv_1208|M.gilvum_PYR-GCK GYVIQSAGLMPHLRVIDNVATVPVLQGESRRSARKSALGVMERVGLDP-K
Mvan_5600|M.vanbaalenii_PYR-1 GYVIQSAGLMPHLRVVDNVATVPVLRGESRRSARRSALGVLERVGLDP-K
MSMEG_6333|M.smegmatis_MC2_155 GYVIQSAGLMPHLRVVDNVATVPVLRGESRRSARKAAIGVMERVGLDP-K
MAB_0262|M.abscessus_ATCC_1997 GYVIQNAGLMPHQRVIDNVATVPVLRGESRRAARKTALEVLERVGLDP-K
TH_4520|M.thermoresistible__bu GYVIQGAGLFPHYTVAENIAIVPRLLKWDRKRIAARVDELLDLVSLDPDE
MUL_0033|M.ulcerans_Agy99 GYVVQGAGVFLHLTVGDNIGIVPRLLKWDKNRVAARIDELLDLVNLDPAQ
MLBr_01089|M.leprae_Br4923 AMVFQSYALYPHMTVRQNIAFPLMLAKVKKAEIAQKVSETAQILDLTD--
. *.* .: * * :*:. * : : :.*
Mb3784c|M.bovis_AF2122/97 VATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHELQNE
Rv3758c|M.tuberculosis_H37Rv VATRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHELQNE
MMAR_5301|M.marinum_M VASRYPAQLSGGEQQRVGVARALAADPPILLMDEAFSAVDPVVRHDLQNE
MAV_0318|M.avium_104 LARRYPAQLSGGEQQRVGVARALAADPPILLMDEPFSAVDPVVRHDLQNE
Mflv_1208|M.gilvum_PYR-GCK LADRYPAQLSGGQQQRVGVARALAADPPILLMDEPFSAVDPVVRDDLQAE
Mvan_5600|M.vanbaalenii_PYR-1 LADRYPAQLSGGQQQRVGVARALAADPPILLMDEPFSAVDPVVREDLQTE
MSMEG_6333|M.smegmatis_MC2_155 LADRYPAQLSGGQQQRVGVARALAADPPILLMDEPFSAVDPVVREDLQAE
MAB_0262|M.abscessus_ATCC_1997 LATRYPAQLSGGQQQRVGVARALAADPPILLMDEPFSAVDPNVRDDLQAE
TH_4520|M.thermoresistible__bu YRDRYPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
MUL_0033|M.ulcerans_Agy99 YRDRFPRELSGGQQQRVGVARALAADPPVLLMDEPFGAVDPITRQRLQDE
MLBr_01089|M.leprae_Br4923 LLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAKLRVRTRGE
* * :****::***.:.**:. .* :****.:. :*. * : *
Mb3784c|M.bovis_AF2122/97 ILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETARLLSS
Rv3758c|M.tuberculosis_H37Rv ILRLQAELHKTIVFVTHDIDEALKLADLVAVFAPGGALAQYDETARLLSS
MMAR_5301|M.marinum_M ILRLQSDLHKTIVFVTHDIDEAFKLGEVVAVFAPGGRLQQCDQPARLLAR
MAV_0318|M.avium_104 ILRLQSELHKTIVFVTHDIDEALRLGERVAVFK-GGMLQQCDEPARLLSR
Mflv_1208|M.gilvum_PYR-GCK IVRLQSELRKTIVFVTHDIDEAIKLGDRVAVFGRGGVLEQYDAPARLLSN
Mvan_5600|M.vanbaalenii_PYR-1 ILRLQSELRKTIVFVTHDIDEAIKLGDKVAVFGRGGVLQQYDAPARLLSN
MSMEG_6333|M.smegmatis_MC2_155 IIRLQGELRKTVIFVTHDIDEAIKLGDKVAVFGRGGVLQQYDAPARLLSN
MAB_0262|M.abscessus_ATCC_1997 MQRLQAELNKTIVFVTHDIDEAVKLGDKVAVFGPGGVLQQYDEPERVLSN
TH_4520|M.thermoresistible__bu LLRLQDELRKTIVFVTHDFDEAVKLGDRIAILQQGSKVVQYDTPEEILTN
MUL_0033|M.ulcerans_Agy99 LLRLQEELHKTIVFVTHDFGEAVKLGDRIAILTTGSKVVQYDTPERVLAE
MLBr_01089|M.leprae_Br4923 IARLQRRLGATTVYVTHDQTEAMTLGDRVVVMR-SGVVQQIGTPDELYER
: *** * * ::**** **. *.: :.:: .. : * . . .:
Mb3784c|M.bovis_AF2122/97 PANDFVSKFIGLGRG--YRWLQLFDAAGLPVRDIEQVSV-----------
Rv3758c|M.tuberculosis_H37Rv PANDFVSKFIGLGRG--YRWLQLFDAAGLPVRDIEQVSV-----------
MMAR_5301|M.marinum_M PANEFVSKFIGLGRG--YRLLQLFDGAGLELHDLERICV-----------
MAV_0318|M.avium_104 PANDFVSRFIGLGRG--YRWLQLIDAAGLPVHDVRRLFA-----------
Mflv_1208|M.gilvum_PYR-GCK PANDSVAGFVGADRG--YRGLQFFHATGLPLHDIRHVGE-----------
Mvan_5600|M.vanbaalenii_PYR-1 PANDEVAEFVGADRG--YRGLQFFHATGLPLHDIRRVAE-----------
MSMEG_6333|M.smegmatis_MC2_155 PANEFVAGFVGADRG--YRGLQFFHATGLPLHEIRHVAE-----------
MAB_0262|M.abscessus_ATCC_1997 PASDFVAGFIGRDRG--YRGLQFREAGEVPLYEIRQVRE-----------
TH_4520|M.thermoresistible__bu PADDFVKGFVGHGAA--LKQLTLTRVRDVDLHDAVTARAG---------G
MUL_0033|M.ulcerans_Agy99 PANDFVRGFVGAGAT--LMQLSLTRVRDIDLHEAVVAQAG---------G
MLBr_01089|M.leprae_Br4923 PVNLFVAGFIGSPVMNFFPAKLTPNGLTLPLGELNLSRDVQAMIGQHPVP
*.. * *:* : : :
Mb3784c|M.bovis_AF2122/97 -NGLSDARDRQVRDGWVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTV
Rv3758c|M.tuberculosis_H37Rv -NGLSDARDRQVRDGWVLVVDGAGAPLGWIDADGRRRHRGGAALSDAMTV
MMAR_5301|M.marinum_M -SSLGRS---QLPDGWALVVSDDDVPVGWIDADGLRRHRDGATLPDAMTV
MAV_0318|M.avium_104 -ENLSGA---DVFDGWAVVVDEAGAPLGWIDAEGLRRHRDGLALSDSMTG
Mflv_1208|M.gilvum_PYR-GCK -PEIDALT--LGSGDWVLVTRPDGTPYAWIDAEGVDLHRKGASLYDSTIA
Mvan_5600|M.vanbaalenii_PYR-1 -PEINALT--LNPGDWVLVTRTDGTPYAWINAAGVELHRNGSSLYDSTIA
MSMEG_6333|M.smegmatis_MC2_155 -PLIDSLD--LAPGDWVLVTKPDGSPYAWINAEGVEVHRSGKSLYDSTIA
MAB_0262|M.abscessus_ATCC_1997 -PQITDLI--IAPGDWVLVCNADGTPFGWMDDAGADVYRSGKSLYDSVIA
TH_4520|M.thermoresistible__bu DPDEAIHAARAADREHVIVLDAQNRPMRWMSLAELARPGALIRVSRDENL
MUL_0033|M.ulcerans_Agy99 DPAEAVELAGSLDRAHAIVLDRQNRPLRWLPLEELAKPQALAQVTRDDDL
MLBr_01089|M.leprae_Br4923 DRVIVGVRPEHLTDAKLIDAHQHGTVLTFKVKVDLVESLGADKYIYFTTT
: . :
Mb3784c|M.bovis_AF2122/97 GGSVFRPNGNLSQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQK
Rv3758c|M.tuberculosis_H37Rv GGSVFRPNGNLSQALDAALSSPSGVGVAVDGGGKVIGGILAADVLAEFQK
MMAR_5301|M.marinum_M VGAVFRPNGNLSQALDAALSSPSSMGVAVDDGGKVIGGVLADDVLSALKS
MAV_0318|M.avium_104 VGSLFRPRGNLSQALDAALSSPSGIGVAVDDDGRVIGGVLAADVLAAVES
Mflv_1208|M.gilvum_PYR-GCK GGSLFRPDGTLRLALDAALSSPSGLGVAVDADGQVVGGVKADDVLAALDT
Mvan_5600|M.vanbaalenii_PYR-1 GGSLFQPDGTLRLALDAALSSPAGLGVAVDGAGQVIGGVRADDVLAALDK
MSMEG_6333|M.smegmatis_MC2_155 GGSLFRPDGTLRHALDAALSSPSGLGVAVDADDHVIGGVRADDVLAALAE
MAB_0262|M.abscessus_ATCC_1997 GGSLFGPDGTLRQALDAALSSPCGLGVAIDADGRVRGGVRGDDVLTALDR
TH_4520|M.thermoresistible__bu ECVTLAS--TLTDALDDMLTSSHGVVVVTGRRNTYQGVIRLETIMDAIAD
MUL_0033|M.ulcerans_Agy99 ECVNLAS--TLNDALDEMLTSAHGVVVVTGRRNTYQGVIRVETIMEAIAD
MLBr_01089|M.leprae_Br4923 GCDVYSAQLDELASELEVRENQFVARVSAESKMAIGESIELAFGTAKIAV
. : . * : .
Mb3784c|M.bovis_AF2122/97 GKKAGGGAKPCTT----
Rv3758c|M.tuberculosis_H37Rv GKKAGGGAKPCTT----
MMAR_5301|M.marinum_M RR---------------
MAV_0318|M.avium_104 QRKSR------------
Mflv_1208|M.gilvum_PYR-GCK QRRARHEG---------
Mvan_5600|M.vanbaalenii_PYR-1 QRAARQAEG--------
MSMEG_6333|M.smegmatis_MC2_155 QRQIPEL----------
MAB_0262|M.abscessus_ATCC_1997 QRRNGQR----------
TH_4520|M.thermoresistible__bu ARGTAGSQGAGAP----
MUL_0033|M.ulcerans_Agy99 IRGAANSEEPDT-----
MLBr_01089|M.leprae_Br4923 FDADSGVNLTVSAPTDA