For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLPYNKNPAAVNALSPEQYHVTQQNGTERPFTGEYWDNHEPGIYVDVVSGEPLFASVDKFESGSGWPSFT RPIDSGNVVEKRDFSHLMIRTEVRSAHGDSHLGHVFKDGPREAGGLRYCINSAALRFVHLDDLEEQGYGQ YRGLFVAETATEEEQA*
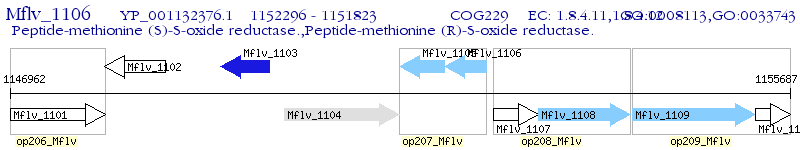
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1106 | - | - | 100% (157) | methionine-R-sulfoxide reductase |
| M. gilvum PYR-GCK | Mflv_3916 | - | 3e-24 | 50.00% (116) | methionine-R-sulfoxide reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2693 | - | 3e-25 | 51.72% (116) | hypothetical protein Mb2693 |
| M. tuberculosis H37Rv | Rv2674 | - | 3e-25 | 51.72% (116) | hypothetical protein Rv2674 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2979 | - | 6e-22 | 47.37% (114) | hypothetical protein MAB_2979 |
| M. marinum M | MMAR_2042 | - | 1e-23 | 50.42% (119) | hypothetical protein MMAR_2042 |
| M. avium 104 | MAV_3566 | msrB | 1e-24 | 50.86% (116) | methionine-R-sulfoxide reductase |
| M. smegmatis MC2 155 | MSMEG_4418 | msrB | 3e-64 | 76.76% (142) | methionine-R-sulfoxide reductase |
| M. thermoresistible (build 8) | TH_0623 | - | 2e-26 | 51.67% (120) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3308 | - | 3e-23 | 49.58% (119) | hypothetical protein MUL_3308 |
| M. vanbaalenii PYR-1 | Mvan_5708 | - | 2e-79 | 85.99% (157) | methionine-R-sulfoxide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2979|M.abscessus_ATCC_1997 MTTSN----DPKVNLTDEQWRERLTPDEFAVLRRAGTERPFVGEYTDTKT
MAV_3566|M.avium_104 MTS-------PKVQLTDDEWRQRLTPEEFHVLRQAGTERPFTGEYTDTKT
TH_0623|M.thermoresistible__bu MTIP-----EPKVQLSDDEWRERLTPEEYYVLRQAGTERPFTGEYTDTKT
Mb2693|M.bovis_AF2122/97 -------MTRPKLELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTT
Rv2674|M.tuberculosis_H37Rv -------MTRPKLELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTT
MMAR_2042|M.marinum_M MTSSDAGSQRPKLQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKT
MUL_3308|M.ulcerans_Agy99 MISSDAGSQRPKLQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKT
Mflv_1106|M.gilvum_PYR-GCK ---------MTLPYNKNPAAVNALSPEQYHVTQQNGTERPFTGEYWDNHE
Mvan_5708|M.vanbaalenii_PYR-1 ---------MTQRYNKNPAAVDALSPEQYHVTQRNGTERPFTGEYWDNHE
MSMEG_4418|M.smegmatis_MC2_155 ----------MHEYRKNPDALEALSPEQYHVTQESGTEPPFSGEYVNNHE
.: . *: :: * : *** ** *** :.
MAB_2979|M.abscessus_ATCC_1997 EGVYNCRACGAELFRSTEKFESHCGWPSFFDPANSDAVILRPDDSLGMRR
MAV_3566|M.avium_104 EGVYQCRACGAELFRSTEKFESHCGWPSFYDPANSDAVILLPDHSLGTTR
TH_0623|M.thermoresistible__bu EGVYTCRACGAELFRSAEKFESHCGWPSFFDPASSDAVITRPDDSLGMRR
Mb2693|M.bovis_AF2122/97 AGIYQCRACGAELFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTR
Rv2674|M.tuberculosis_H37Rv AGIYQCRACGAELFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTR
MMAR_2042|M.marinum_M QGIYQCRACGAELFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLR
MUL_3308|M.ulcerans_Agy99 QGIYQCRACGAELFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLR
Mflv_1106|M.gilvum_PYR-GCK PGIYVDVVSGEPLFASVDKFESGSGWPSFTRPIDSGNVVEKRDFSHLMIR
Mvan_5708|M.vanbaalenii_PYR-1 PGLYVDVVSGEPLFASVDKFDSGSGWPSFTRPIEKENVLEKRDFSHLMIR
MSMEG_4418|M.smegmatis_MC2_155 PGIYVDVVSGEPLFASADKFDSGTGWPSFTRPLDDGYVVEKSDRSHFMVR
*:* ..* ** *.:**:* ***** * . * * * *
MAB_2979|M.abscessus_ATCC_1997 VEVLCANCHSHLGHVFEGEGYPTPTDQRYCINSISLRLAPQ---------
MAV_3566|M.avium_104 TEVLCANCHSHLGHVFTGEGYPTPTDQRYCINSICLRLVPS---------
TH_0623|M.thermoresistible__bu TEVLCASCHSHLGHVFEGEGYPTPTDLRYCINSISLRLVPAAD-------
Mb2693|M.bovis_AF2122/97 TEVLCANCDSHLGHVFAGEGYPTPTDKRYCINSISLRLVPGSV-------
Rv2674|M.tuberculosis_H37Rv TEVLCANCDSHLGHVFAGEGYPTPTDKRYCINSISLRLVPGSV-------
MMAR_2042|M.marinum_M TEVLCANCHSHLGHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ----
MUL_3308|M.ulcerans_Agy99 TEVLCANCHGHLGHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ----
Mflv_1106|M.gilvum_PYR-GCK TEVRSAHGDSHLGHVFK-DGPREAGGLRYCINSAALRFVHLDDLEEQGYG
Mvan_5708|M.vanbaalenii_PYR-1 TEVRSAHGDSHLGHVFN-DGPRDRGGLRYCVNSAALRFIHLDDLEAQGYG
MSMEG_4418|M.smegmatis_MC2_155 TEVRSRHGDSHLGHVFT-DGPMSDGGLRYCINSASLRFIHRDDLEAEGYG
.** . ..****** :* . ***:** .**:
MAB_2979|M.abscessus_ATCC_1997 -------------------------
MAV_3566|M.avium_104 -------------------------
TH_0623|M.thermoresistible__bu -------------------------
Mb2693|M.bovis_AF2122/97 -------------------------
Rv2674|M.tuberculosis_H37Rv -------------------------
MMAR_2042|M.marinum_M -------------------------
MUL_3308|M.ulcerans_Agy99 -------------------------
Mflv_1106|M.gilvum_PYR-GCK QYRGLFVAE-TATEEEQA-------
Mvan_5708|M.vanbaalenii_PYR-1 QYRGLF----EATDKEQA-------
MSMEG_4418|M.smegmatis_MC2_155 RYLSLFDSSGDSTRHTDSQLNEVQP