For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSPKVQLTDDEWRQRLTPEEFHVLRQAGTERPFTGEYTDTKTEGVYQCRACGAELFRSTEKFESHCGWP SFYDPANSDAVILLPDHSLGTTRTEVLCANCHSHLGHVFTGEGYPTPTDQRYCINSICLRLVPS
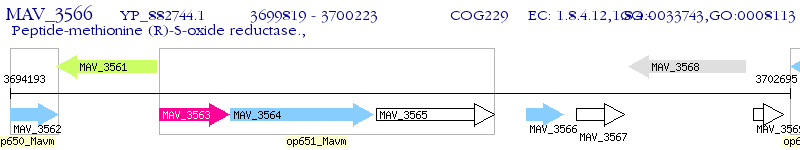
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3566 | msrB | - | 100% (134) | methionine-R-sulfoxide reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2693 | - | 5e-69 | 84.96% (133) | hypothetical protein Mb2693 |
| M. gilvum PYR-GCK | Mflv_3916 | - | 1e-68 | 85.82% (134) | methionine-R-sulfoxide reductase |
| M. tuberculosis H37Rv | Rv2674 | - | 5e-69 | 84.96% (133) | hypothetical protein Rv2674 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2979 | - | 3e-67 | 86.15% (130) | hypothetical protein MAB_2979 |
| M. marinum M | MMAR_2042 | - | 2e-65 | 82.44% (131) | hypothetical protein MMAR_2042 |
| M. smegmatis MC2 155 | MSMEG_2784 | msrB | 1e-67 | 84.85% (132) | methionine-R-sulfoxide reductase |
| M. thermoresistible (build 8) | TH_0623 | - | 1e-67 | 86.26% (131) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3308 | - | 5e-65 | 81.68% (131) | hypothetical protein MUL_3308 |
| M. vanbaalenii PYR-1 | Mvan_2487 | - | 1e-68 | 85.07% (134) | methionine-R-sulfoxide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2693|M.bovis_AF2122/97 MTR-------PKLELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTT
Rv2674|M.tuberculosis_H37Rv MTR-------PKLELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTT
Mflv_3916|M.gilvum_PYR-GCK MTA-------PKLVLSDDQWRERLTPQEFAVLRQAGTERPFTGEYTDTKT
Mvan_2487|M.vanbaalenii_PYR-1 MTG-------PKLVLSDDQWRERLTPQEFAVLRQAGTERPFTGEYTDTKT
MAB_2979|M.abscessus_ATCC_1997 MTTSND----PKVNLTDEQWRERLTPDEFAVLRRAGTERPFVGEYTDTKT
MSMEG_2784|M.smegmatis_MC2_155 MTIP-A----PKLSLTDDEWREKLTPQEFAVLRRAGTERPFTGEYTDTKT
TH_0623|M.thermoresistible__bu MTIP-E----PKVQLSDDEWRERLTPEEYYVLRQAGTERPFTGEYTDTKT
MAV_3566|M.avium_104 MTS-------PKVQLTDDEWRQRLTPEEFHVLRQAGTERPFTGEYTDTKT
MMAR_2042|M.marinum_M MTSSDAGSQRPKLQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKT
MUL_3308|M.ulcerans_Agy99 MISSDAGSQRPKLQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKT
* **: *::::**::*: *: *** *******.******.*
Mb2693|M.bovis_AF2122/97 AGIYQCRACGAELFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTR
Rv2674|M.tuberculosis_H37Rv AGIYQCRACGAELFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTR
Mflv_3916|M.gilvum_PYR-GCK QGVYQCRACGAELFRSTEKFESHCGWPSFFDPADSDAVILRTDDSLGMRR
Mvan_2487|M.vanbaalenii_PYR-1 DGVYQCRACGAELFRSSEKFESHCGWPSFFDPADSDAVLLRPDDSLGMRR
MAB_2979|M.abscessus_ATCC_1997 EGVYNCRACGAELFRSTEKFESHCGWPSFFDPANSDAVILRPDDSLGMRR
MSMEG_2784|M.smegmatis_MC2_155 EGVYQCRACGAELFRSSEKFESHCGWPSFFDPADSDAVILRPDDSLGMRR
TH_0623|M.thermoresistible__bu EGVYTCRACGAELFRSAEKFESHCGWPSFFDPASSDAVITRPDDSLGMRR
MAV_3566|M.avium_104 EGVYQCRACGAELFRSTEKFESHCGWPSFYDPANSDAVILLPDHSLGTTR
MMAR_2042|M.marinum_M QGIYQCRACGAELFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLR
MUL_3308|M.ulcerans_Agy99 QGIYQCRACGAELFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLR
*:* ***********:************:** *:** .*.*** *
Mb2693|M.bovis_AF2122/97 TEVLCANCDSHLGHVFAGEGYPTPTDKRYCINSISLRLVPGSV---
Rv2674|M.tuberculosis_H37Rv TEVLCANCDSHLGHVFAGEGYPTPTDKRYCINSISLRLVPGSV---
Mflv_3916|M.gilvum_PYR-GCK VEVLCANCHSHLGHVFEGEGYPTPTDQRYCINSISLRLVPSGS---
Mvan_2487|M.vanbaalenii_PYR-1 VEVLCANCHSHLGHVFEGEGYPTPTDQRYCINSISLRLVPSQS---
MAB_2979|M.abscessus_ATCC_1997 VEVLCANCHSHLGHVFEGEGYPTPTDQRYCINSISLRLAPQ-----
MSMEG_2784|M.smegmatis_MC2_155 VEVLCANCHSHLGHVFEGEGYPTPTDKRYCINSISLRLVPAE----
TH_0623|M.thermoresistible__bu TEVLCASCHSHLGHVFEGEGYPTPTDLRYCINSISLRLVPAAD---
MAV_3566|M.avium_104 TEVLCANCHSHLGHVFTGEGYPTPTDQRYCINSICLRLVPS-----
MMAR_2042|M.marinum_M TEVLCANCHSHLGHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ
MUL_3308|M.ulcerans_Agy99 TEVLCANCHGHLGHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ
.*****.*..****** ********* *******.***.*