For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HEYRKNPDALEALSPEQYHVTQESGTEPPFSGEYVNNHEPGIYVDVVSGEPLFASADKFDSGTGWPSFTR PLDDGYVVEKSDRSHFMVRTEVRSRHGDSHLGHVFTDGPMSDGGLRYCINSASLRFIHRDDLEAEGYGRY LSLFDSSGDSTRHTDSQLNEVQP*
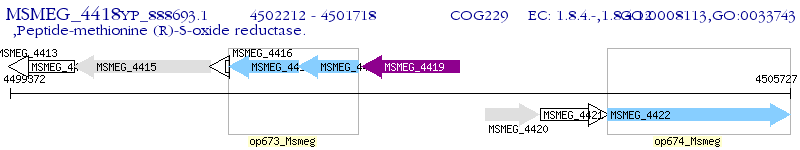
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4418 | msrB | - | 100% (164) | methionine-R-sulfoxide reductase |
| M. smegmatis MC2 155 | MSMEG_2784 | msrB | 6e-22 | 44.07% (118) | methionine-R-sulfoxide reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2693 | - | 1e-23 | 45.76% (118) | hypothetical protein Mb2693 |
| M. gilvum PYR-GCK | Mflv_1106 | - | 2e-64 | 76.76% (142) | methionine-R-sulfoxide reductase |
| M. tuberculosis H37Rv | Rv2674 | - | 1e-23 | 45.76% (118) | hypothetical protein Rv2674 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1050c | - | 4e-24 | 47.41% (116) | putative methionine sulfoxide reductase |
| M. marinum M | MMAR_2042 | - | 1e-21 | 45.38% (119) | hypothetical protein MMAR_2042 |
| M. avium 104 | MAV_3566 | msrB | 7e-23 | 44.07% (118) | methionine-R-sulfoxide reductase |
| M. thermoresistible (build 8) | TH_0623 | - | 7e-25 | 46.72% (122) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3308 | - | 3e-21 | 44.54% (119) | hypothetical protein MUL_3308 |
| M. vanbaalenii PYR-1 | Mvan_5708 | - | 8e-64 | 73.79% (145) | methionine-R-sulfoxide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2693|M.bovis_AF2122/97 --------------------------------------MT-------RPK
Rv2674|M.tuberculosis_H37Rv --------------------------------------MT-------RPK
MMAR_2042|M.marinum_M --------------------------------------MTSSDAGSQRPK
MUL_3308|M.ulcerans_Agy99 --------------------------------------MISSDAGSQRPK
MAV_3566|M.avium_104 --------------------------------------MT-------SPK
TH_0623|M.thermoresistible__bu --------------------------------------MTIP-----EPK
MAB_1050c|M.abscessus_ATCC_199 MRRWSPSAADPALTRRQLLVGVVAVCALACLDRYDSAAADPSTPSAPVPS
Mflv_1106|M.gilvum_PYR-GCK -----------------------------------------------MTL
Mvan_5708|M.vanbaalenii_PYR-1 -----------------------------------------------MTQ
MSMEG_4418|M.smegmatis_MC2_155 ------------------------------------------------MH
Mb2693|M.bovis_AF2122/97 LELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTTAGIYQCRACGAE
Rv2674|M.tuberculosis_H37Rv LELSDDEWRQKLTPQEFHVLRRAGTERPFTGEYTDTTTAGIYQCRACGAE
MMAR_2042|M.marinum_M LQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKTQGIYQCRACGAE
MUL_3308|M.ulcerans_Agy99 LQLSEDEWRKRLSQREYDVLRLAGTERPFTGEYTDTKTQGIYQCRACGAE
MAV_3566|M.avium_104 VQLTDDEWRQRLTPEEFHVLRQAGTERPFTGEYTDTKTEGVYQCRACGAE
TH_0623|M.thermoresistible__bu VQLSDDEWRERLTPEEYYVLRQAGTERPFTGEYTDTKTEGVYTCRACGAE
MAB_1050c|M.abscessus_ATCC_199 IVHTDAEWRQLLTPSQYQILRKAGTETPYSSPLNNEHRTGVFSCAGCALD
Mflv_1106|M.gilvum_PYR-GCK PYNKNPAAVNALSPEQYHVTQQNGTERPFTGEYWDNHEPGIYVDVVSGEP
Mvan_5708|M.vanbaalenii_PYR-1 RYNKNPAAVDALSPEQYHVTQRNGTERPFTGEYWDNHEPGLYVDVVSGEP
MSMEG_4418|M.smegmatis_MC2_155 EYRKNPDALEALSPEQYHVTQESGTEPPFSGEYVNNHEPGIYVDVVSGEP
.: . *: :: : : *** *::. : *:: ..
Mb2693|M.bovis_AF2122/97 LFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTRTEVLCANCDSHL
Rv2674|M.tuberculosis_H37Rv LFRSTEKFESHCGWPSFFDPKSSDAVTLRPDHSLGMTRTEVLCANCDSHL
MMAR_2042|M.marinum_M LFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLRTEVLCANCHSHL
MUL_3308|M.ulcerans_Agy99 LFRSTEKFESHCGWPSFFDPSASEAVILRPDHSLGQLRTEVLCANCHGHL
MAV_3566|M.avium_104 LFRSTEKFESHCGWPSFYDPANSDAVILLPDHSLGTTRTEVLCANCHSHL
TH_0623|M.thermoresistible__bu LFRSAEKFESHCGWPSFFDPASSDAVITRPDDSLGMRRTEVLCASCHSHL
MAB_1050c|M.abscessus_ATCC_199 LFSSAAKFDSGTGWPSFWQP-LPHAVSERPDNSLGMSRTEVFCSRCAGHL
Mflv_1106|M.gilvum_PYR-GCK LFASVDKFESGSGWPSFTRPIDSGNVVEKRDFSHLMIRTEVRSAHGDSHL
Mvan_5708|M.vanbaalenii_PYR-1 LFASVDKFDSGSGWPSFTRPIEKENVLEKRDFSHLMIRTEVRSAHGDSHL
MSMEG_4418|M.smegmatis_MC2_155 LFASADKFDSGTGWPSFTRPLDDGYVVEKSDRSHFMVRTEVRSRHGDSHL
** *. **:* ***** * * * * **** . .**
Mb2693|M.bovis_AF2122/97 GHVFAGEGYPTPTDKRYCINSISLRLVPGSV-------------------
Rv2674|M.tuberculosis_H37Rv GHVFAGEGYPTPTDKRYCINSISLRLVPGSV-------------------
MMAR_2042|M.marinum_M GHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ----------------
MUL_3308|M.ulcerans_Agy99 GHVFAGEGYPTPTDLRYCINSISLRLVPTDSAAQ----------------
MAV_3566|M.avium_104 GHVFTGEGYPTPTDQRYCINSICLRLVPS---------------------
TH_0623|M.thermoresistible__bu GHVFEGEGYPTPTDLRYCINSISLRLVPAAD-------------------
MAB_1050c|M.abscessus_ATCC_199 GHVFD-DG-PKPTGLRYCMNGAAMNFRPT---------------------
Mflv_1106|M.gilvum_PYR-GCK GHVFK-DGPREAGGLRYCINSAALRFVHLDDLEEQGYGQYRGLFVAE-TA
Mvan_5708|M.vanbaalenii_PYR-1 GHVFN-DGPRDRGGLRYCVNSAALRFIHLDDLEAQGYGQYRGLF----EA
MSMEG_4418|M.smegmatis_MC2_155 GHVFT-DGPMSDGGLRYCINSASLRFIHRDDLEAEGYGRYLSLFDSSGDS
**** :* . ***:*. .:.:
Mb2693|M.bovis_AF2122/97 -------------
Rv2674|M.tuberculosis_H37Rv -------------
MMAR_2042|M.marinum_M -------------
MUL_3308|M.ulcerans_Agy99 -------------
MAV_3566|M.avium_104 -------------
TH_0623|M.thermoresistible__bu -------------
MAB_1050c|M.abscessus_ATCC_199 -------------
Mflv_1106|M.gilvum_PYR-GCK TEEEQA-------
Mvan_5708|M.vanbaalenii_PYR-1 TDKEQA-------
MSMEG_4418|M.smegmatis_MC2_155 TRHTDSQLNEVQP