For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPVDDADHCPASGSVHHGYITDKDKYLKRLKRIEGQARGISRMIEEERYCIDILTQADALTKALQSVSL ALLDDHLRHCVRDAAAAGGPAADAKLTEASEAIARLVRS
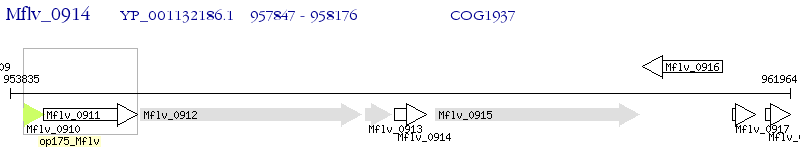
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0914 | - | - | 100% (109) | hypothetical protein Mflv_0914 |
| M. gilvum PYR-GCK | Mflv_2851 | - | 3e-57 | 98.17% (109) | hypothetical protein Mflv_2851 |
| M. gilvum PYR-GCK | Mflv_0484 | - | 2e-31 | 67.39% (92) | hypothetical protein Mflv_0484 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0196 | - | 1e-32 | 68.48% (92) | hypothetical protein Mb0196 |
| M. tuberculosis H37Rv | Rv0190 | - | 2e-32 | 68.48% (92) | hypothetical protein Rv0190 |
| M. leprae Br4923 | MLBr_02609 | - | 7e-28 | 61.96% (92) | hypothetical protein MLBr_02609 |
| M. abscessus ATCC 19977 | MAB_4542c | - | 8e-30 | 64.21% (95) | hypothetical protein MAB_4542c |
| M. marinum M | MMAR_0433 | - | 6e-30 | 65.22% (92) | hypothetical protein MMAR_0433 |
| M. avium 104 | MAV_4988 | - | 1e-30 | 67.39% (92) | hypothetical protein MAV_4988 |
| M. smegmatis MC2 155 | MSMEG_0230 | - | 2e-30 | 65.22% (92) | hypothetical protein MSMEG_0230 |
| M. thermoresistible (build 8) | TH_0982 | - | 1e-31 | 68.48% (92) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1083 | - | 4e-30 | 65.22% (92) | hypothetical protein MUL_1083 |
| M. vanbaalenii PYR-1 | Mvan_0175 | - | 2e-30 | 59.81% (107) | hypothetical protein Mvan_0175 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0433|M.marinum_M -----------------------------------MTTA-----------
MUL_1083|M.ulcerans_Agy99 -----------------------------------MTTA-----------
MLBr_02609|M.leprae_Br4923 ----------------MLDHRRSRRSWQPRSIVSQMLSAVLGWLGEGLNE
MAV_4988|M.avium_104 -----------------------------------MTNE-----------
Mb0196|M.bovis_AF2122/97 -----------------------------------MTAA-----------
Rv0190|M.tuberculosis_H37Rv -----------------------------------MTAA-----------
MSMEG_0230|M.smegmatis_MC2_155 -----------------------------------MDATD----------
TH_0982|M.thermoresistible__bu -----------MTQAKPDGA---KPEGA---KPGGAKPDG----------
Mvan_0175|M.vanbaalenii_PYR-1 MSGANSQAPGGANSQAPGGANSQAPGGANSQAPGGANSQGP------TEN
MAB_4542c|M.abscessus_ATCC_199 -----------------------------------MSTKT----------
Mflv_0914|M.gilvum_PYR-GCK --------------------------MTPVDDADHCPASG----------
MMAR_0433|M.marinum_M ---------HGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQI
MUL_1083|M.ulcerans_Agy99 ---------HGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQI
MLBr_02609|M.leprae_Br4923 DTVREDDNRYGYSQQKGNYAKRLRRIEGQVRGIARMIDEDKYCIDILTQI
MAV_4988|M.avium_104 ---------HGYSQQKDNYAKRLRRIEGQVRGIARMIEEDKYCIDVLTQI
Mb0196|M.bovis_AF2122/97 ---------HGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQI
Rv0190|M.tuberculosis_H37Rv ---------HGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQI
MSMEG_0230|M.smegmatis_MC2_155 -----ESGVHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQI
TH_0982|M.thermoresistible__bu -----HAGAHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQI
Mvan_0175|M.vanbaalenii_PYR-1 APEAVAADAHGYSGQKANYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQI
MAB_4542c|M.abscessus_ATCC_199 ------STGHGYSLKKDDYAKRLLRIEGQVRGIAKMIDEDKYCIDVLTQI
Mflv_0914|M.gilvum_PYR-GCK ------SVHHGYITDKDKYLKRLKRIEGQARGISRMIEEERYCIDILTQA
:** .* .* *** *:***.***::**:*::****:***
MMAR_0433|M.marinum_M SAVNSALRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVR
MUL_1083|M.ulcerans_Agy99 SAVNSALRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVR
MLBr_02609|M.leprae_Br4923 SAVSNALRSVALNLLDEHLEYCVSRAVAEGGSEAEDKFAEASAAIARLVR
MAV_4988|M.avium_104 SAVNSALRSVALNLLDEHLGHCVTRAVAGGGDDADEKLAEASAAIARLVR
Mb0196|M.bovis_AF2122/97 SAVTSALRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVR
Rv0190|M.tuberculosis_H37Rv SAVTSALRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVR
MSMEG_0230|M.smegmatis_MC2_155 SAVNSALQSVALGLLDEHLGHCVSQAVAEGGQEAEAKLAEASAAIARLVR
TH_0982|M.thermoresistible__bu SAANSALQSVALALLDEHLGHCVTQAVAEGGEAAEAKLAEASAAIARLVR
Mvan_0175|M.vanbaalenii_PYR-1 SAVNSALQSVALGLLDEHLSHCVSHAVTVGGSEADAKLAEASAAIARLVR
MAB_4542c|M.abscessus_ATCC_199 SAAQSALRSVALGLLDEHLGHCVTNAVASGGADADEKLAEASAAIARLVR
Mflv_0914|M.gilvum_PYR-GCK DALTKALQSVSLALLDDHLRHCVRDAAAAGGPAADAKLTEASEAIARLVR
.* .**:**:* ***:** :** *.: ** *: *::*** *******
MMAR_0433|M.marinum_M S
MUL_1083|M.ulcerans_Agy99 S
MLBr_02609|M.leprae_Br4923 S
MAV_4988|M.avium_104 S
Mb0196|M.bovis_AF2122/97 S
Rv0190|M.tuberculosis_H37Rv S
MSMEG_0230|M.smegmatis_MC2_155 S
TH_0982|M.thermoresistible__bu S
Mvan_0175|M.vanbaalenii_PYR-1 S
MAB_4542c|M.abscessus_ATCC_199 S
Mflv_0914|M.gilvum_PYR-GCK S
*