For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDATDESGVHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQISAVNSALQSVALGLLDEHLG HCVSQAVAEGGQEAEAKLAEASAAIARLVRS
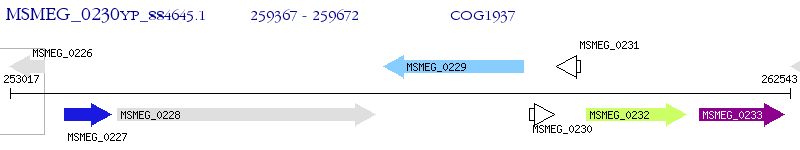
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0230 | - | - | 100% (101) | hypothetical protein MSMEG_0230 |
| M. smegmatis MC2 155 | MSMEG_5388 | - | 9e-09 | 36.84% (76) | hypothetical protein MSMEG_5388 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0196 | - | 3e-39 | 80.00% (95) | hypothetical protein Mb0196 |
| M. gilvum PYR-GCK | Mflv_0484 | - | 2e-44 | 89.80% (98) | hypothetical protein Mflv_0484 |
| M. tuberculosis H37Rv | Rv0190 | - | 3e-39 | 80.00% (95) | hypothetical protein Rv0190 |
| M. leprae Br4923 | MLBr_02609 | - | 6e-39 | 79.38% (97) | hypothetical protein MLBr_02609 |
| M. abscessus ATCC 19977 | MAB_4542c | - | 2e-39 | 79.80% (99) | hypothetical protein MAB_4542c |
| M. marinum M | MMAR_0433 | - | 2e-41 | 88.04% (92) | hypothetical protein MMAR_0433 |
| M. avium 104 | MAV_4988 | - | 2e-40 | 85.87% (92) | hypothetical protein MAV_4988 |
| M. thermoresistible (build 8) | TH_0982 | - | 4e-45 | 91.58% (95) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1083 | - | 2e-41 | 88.04% (92) | hypothetical protein MUL_1083 |
| M. vanbaalenii PYR-1 | Mvan_0175 | - | 4e-42 | 90.22% (92) | hypothetical protein Mvan_0175 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0196|M.bovis_AF2122/97 MT------------------------------------------------
Rv0190|M.tuberculosis_H37Rv MT------------------------------------------------
MSMEG_0230|M.smegmatis_MC2_155 ---------------------------MD-----------ATDES-----
TH_0982|M.thermoresistible__bu MTQAK------PDGAKPEGAK---PGGAK-----------PDGHA-----
Mflv_0484|M.gilvum_PYR-GCK MS---------DDSTTP-----------------------PDE-------
Mvan_0175|M.vanbaalenii_PYR-1 MSGANSQAPGGANSQAPGGANSQAPGGANSQAPGGANSQGPTENAPEAVA
MAB_4542c|M.abscessus_ATCC_199 MS---------------------------------------TKTS-----
MMAR_0433|M.marinum_M MT------------------------------------------------
MUL_1083|M.ulcerans_Agy99 MT------------------------------------------------
MAV_4988|M.avium_104 MT------------------------------------------------
MLBr_02609|M.leprae_Br4923 MLDHRRSRRSWQPRSIVSQMLSAVLGWLG----------EGLNEDTVRED
Mb0196|M.bovis_AF2122/97 -AAHGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQISAVTSA
Rv0190|M.tuberculosis_H37Rv -AAHGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQISAVTSA
MSMEG_0230|M.smegmatis_MC2_155 -GVHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQISAVNSA
TH_0982|M.thermoresistible__bu -GAHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQISAANSA
Mflv_0484|M.gilvum_PYR-GCK --THGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQISAVNSA
Mvan_0175|M.vanbaalenii_PYR-1 ADAHGYSGQKANYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQISAVNSA
MAB_4542c|M.abscessus_ATCC_199 -TGHGYSLKKDDYAKRLLRIEGQVRGIAKMIDEDKYCIDVLTQISAAQSA
MMAR_0433|M.marinum_M -TAHGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQISAVNSA
MUL_1083|M.ulcerans_Agy99 -TAHGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQISAVNSA
MAV_4988|M.avium_104 -NEHGYSQQKDNYAKRLRRIEGQVRGIARMIEEDKYCIDVLTQISAVNSA
MLBr_02609|M.leprae_Br4923 DNRYGYSQQKGNYAKRLRRIEGQVRGIARMIDEDKYCIDILTQISAVSNA
:**: :* :***** *:********:**:*******:******. .*
Mb0196|M.bovis_AF2122/97 LRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVRS
Rv0190|M.tuberculosis_H37Rv LRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVRS
MSMEG_0230|M.smegmatis_MC2_155 LQSVALGLLDEHLGHCVSQAVAEGGQEAEAKLAEASAAIARLVRS
TH_0982|M.thermoresistible__bu LQSVALALLDEHLGHCVTQAVAEGGEAAEAKLAEASAAIARLVRS
Mflv_0484|M.gilvum_PYR-GCK LQSVALGLLDEHLGHCVSHAVAAGGADADAKLAEASAAIARLVRS
Mvan_0175|M.vanbaalenii_PYR-1 LQSVALGLLDEHLSHCVSHAVTVGGSEADAKLAEASAAIARLVRS
MAB_4542c|M.abscessus_ATCC_199 LRSVALGLLDEHLGHCVTNAVASGGADADEKLAEASAAIARLVRS
MMAR_0433|M.marinum_M LRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVRS
MUL_1083|M.ulcerans_Agy99 LRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVRS
MAV_4988|M.avium_104 LRSVALNLLDEHLGHCVTRAVAGGGDDADEKLAEASAAIARLVRS
MLBr_02609|M.leprae_Br4923 LRSVALNLLDEHLEYCVSRAVAEGGSEAEDKFAEASAAIARLVRS
*:**** ****** :**:.**: ** *: *:*************