For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAAHGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQISAVTSALRSVALNLLDEHLSHCVTR AVAEGGPGADGKLAEASAAIARLVRS
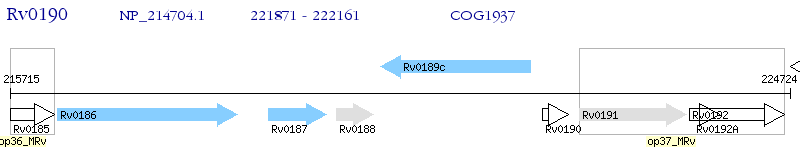
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0190 | - | - | 100% (96) | hypothetical protein Rv0190 |
| M. tuberculosis H37Rv | Rv0967 | - | 2e-11 | 42.50% (80) | hypothetical protein Rv0967 |
| M. tuberculosis H37Rv | Rv1766 | - | 4e-09 | 38.16% (76) | hypothetical protein Rv1766 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0196 | - | 9e-50 | 100.00% (96) | hypothetical protein Mb0196 |
| M. gilvum PYR-GCK | Mflv_0484 | - | 9e-39 | 82.61% (92) | hypothetical protein Mflv_0484 |
| M. leprae Br4923 | MLBr_02609 | - | 2e-39 | 82.61% (92) | hypothetical protein MLBr_02609 |
| M. abscessus ATCC 19977 | MAB_4542c | - | 1e-37 | 78.95% (95) | hypothetical protein MAB_4542c |
| M. marinum M | MMAR_0433 | - | 4e-45 | 90.62% (96) | hypothetical protein MMAR_0433 |
| M. avium 104 | MAV_4988 | - | 2e-43 | 89.58% (96) | hypothetical protein MAV_4988 |
| M. smegmatis MC2 155 | MSMEG_0230 | - | 5e-39 | 80.00% (95) | hypothetical protein MSMEG_0230 |
| M. thermoresistible (build 8) | TH_0982 | - | 2e-39 | 81.72% (93) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1083 | - | 3e-45 | 90.62% (96) | hypothetical protein MUL_1083 |
| M. vanbaalenii PYR-1 | Mvan_0175 | - | 3e-38 | 81.72% (93) | hypothetical protein Mvan_0175 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0190|M.tuberculosis_H37Rv ------------------------------------------MTA-----
Mb0196|M.bovis_AF2122/97 ------------------------------------------MTA-----
Mflv_0484|M.gilvum_PYR-GCK MS---------DDSTTP-----------------------PDE-------
Mvan_0175|M.vanbaalenii_PYR-1 MSGANSQAPGGANSQAPGGANSQAPGGANSQAPGGANSQGPTENAPEAVA
MSMEG_0230|M.smegmatis_MC2_155 --------------------------------------MDATDES-----
TH_0982|M.thermoresistible__bu MTQAKPDGAKPEGAKPGG--------------------AKPDGHA-----
MAB_4542c|M.abscessus_ATCC_199 ---------------------------------------MSTKTS-----
MMAR_0433|M.marinum_M ------------------------------------------MTT-----
MUL_1083|M.ulcerans_Agy99 ------------------------------------------MTT-----
MAV_4988|M.avium_104 ------------------------------------------MTN-----
MLBr_02609|M.leprae_Br4923 ----MLDHRRSRRSWQPRSIVSQMLSAVLGWLGEGLNEDTVREDD-----
Rv0190|M.tuberculosis_H37Rv --AHGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQISAVTSA
Mb0196|M.bovis_AF2122/97 --AHGYTQQKDNYAKRLRRVEGQVRGIARMIEEDKYCIDVLTQISAVTSA
Mflv_0484|M.gilvum_PYR-GCK --THGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQISAVNSA
Mvan_0175|M.vanbaalenii_PYR-1 ADAHGYSGQKANYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQISAVNSA
MSMEG_0230|M.smegmatis_MC2_155 -GVHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDVLTQISAVNSA
TH_0982|M.thermoresistible__bu -GAHGYSAQKENYAKRLRRIEGQVRGIAKMIDEDKYCIDILTQISAANSA
MAB_4542c|M.abscessus_ATCC_199 -TGHGYSLKKDDYAKRLLRIEGQVRGIAKMIDEDKYCIDVLTQISAAQSA
MMAR_0433|M.marinum_M --AHGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQISAVNSA
MUL_1083|M.ulcerans_Agy99 --AHGYSQQKENYAKRLRRIEGQVRGIARMIDEDKYCIDVLTQISAVNSA
MAV_4988|M.avium_104 --EHGYSQQKDNYAKRLRRIEGQVRGIARMIEEDKYCIDVLTQISAVNSA
MLBr_02609|M.leprae_Br4923 -NRYGYSQQKGNYAKRLRRIEGQVRGIARMIDEDKYCIDILTQISAVSNA
:**: :* :***** *:********:**:*******:******. .*
Rv0190|M.tuberculosis_H37Rv LRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVRS
Mb0196|M.bovis_AF2122/97 LRSVALNLLDEHLSHCVTRAVAEGGPGADGKLAEASAAIARLVRS
Mflv_0484|M.gilvum_PYR-GCK LQSVALGLLDEHLGHCVSHAVAAGGADADAKLAEASAAIARLVRS
Mvan_0175|M.vanbaalenii_PYR-1 LQSVALGLLDEHLSHCVSHAVTVGGSEADAKLAEASAAIARLVRS
MSMEG_0230|M.smegmatis_MC2_155 LQSVALGLLDEHLGHCVSQAVAEGGQEAEAKLAEASAAIARLVRS
TH_0982|M.thermoresistible__bu LQSVALALLDEHLGHCVTQAVAEGGEAAEAKLAEASAAIARLVRS
MAB_4542c|M.abscessus_ATCC_199 LRSVALGLLDEHLGHCVTNAVASGGADADEKLAEASAAIARLVRS
MMAR_0433|M.marinum_M LRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVRS
MUL_1083|M.ulcerans_Agy99 LRSVALNLLDEHLNHCVTRAVAEGGTDADGKLAEASAAIARLVRS
MAV_4988|M.avium_104 LRSVALNLLDEHLGHCVTRAVAGGGDDADEKLAEASAAIARLVRS
MLBr_02609|M.leprae_Br4923 LRSVALNLLDEHLEYCVSRAVAEGGSEAEDKFAEASAAIARLVRS
*:**** ****** :**:.**: ** *: *:*************