For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAGAHRAKPLPTRGRSQSRRLLPYTWLGAGAVALGVGAALTGGTAPAHADSGGESSQTHGESRQSRASRD TAERPSQTRSADDGAGSNSDDRSTDDERPLATGNKTRLSKTTRESSSGEARFADAVSEEAHTGAETPAST ETSGLEVRPGDAATDNPDDTAVRAAALAPAARTPSTQDDGTPDAQRTAHVHSGALPAAEGNRASAAGSSL GALFRNQTPTVSWTQASPTGSSVVTGVLTASDPDGDALSYKVTRNPAYGAVTVYADGSVAYTPHQNVATS IDSFRVTVSDAASGFHVHGLSGLINLFSFGLLGKSGHEHTITAEVRFAATAVPQPTVLDRIVLPLNAGQS PRTVAVSPDGRLVFVGSGRWNSPCCGFDNPDGALSVIDAASGAVLSTVKTGLTPISFAVGSTGLLYAVGV DGGTGYTNYFLSAMTPNDYLTKSTFTDTRAIPDLYMQPISLAVTPDGNSVVIGGRNGQVAVVDPMTTRVI RTFDIGFGNHAEGVYVTPDGKYAWISGGYYQPLFRLDLSTGAIGQVQIDYGAGGVLSPDGSKFYTVVDRH DYGQRIEIVDTATGAISSYDYRSESVHRNLIDLAVSPDGKHLYARIAASQTSNRLDVIDAATGVVVAQTQ VGSEAALLGLPGMAISPEGNRIYLATGDSITVVRANSTDRPLLPRAFPSASASALLLNLPRQTDTIYAEK VMRDGKTRMIVYMTGIDAGLNESTLASIISNSGLLHADVEAYIDNAYTAWGGARVIEEIQLVGHSNGGQQ MQAYADWGTYADKVTSVVLFGAPIIKTEDEFTSDALAFINRGDPVPTIFSHWENPGVRWNSMNKDINFYG PESSDALNFERHHASSYYAAAVDFDAKAGLSSAPATQRRLRDTLLRFGGYPVDAMPTAHIAVFTSVTRV
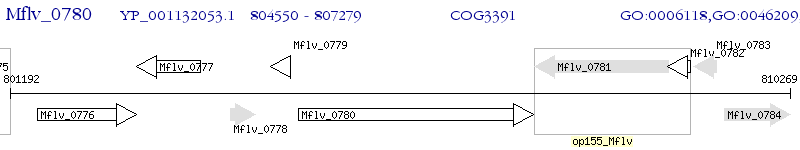
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0780 | - | - | 100% (909) | hypothetical protein Mflv_0780 |
| M. gilvum PYR-GCK | Mflv_5245 | - | 2e-30 | 35.25% (295) | glucose/sorbosone dehydrogenases-like protein |
| M. gilvum PYR-GCK | Mflv_0862 | - | 2e-22 | 26.71% (337) | hypothetical protein Mflv_0862 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0912 | - | 1e-07 | 27.62% (210) | hypothetical protein Mb0912 |
| M. tuberculosis H37Rv | Rv0888 | - | 1e-07 | 27.62% (210) | hypothetical protein Rv0888 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4644 | - | 2e-08 | 25.29% (257) | hypothetical protein MMAR_4644 |
| M. avium 104 | MAV_1185 | - | 2e-06 | 23.56% (208) | hypothetical protein MAV_1185 |
| M. smegmatis MC2 155 | MSMEG_4365 | - | 8e-09 | 26.36% (330) | hypothetical protein MSMEG_4365 |
| M. thermoresistible (build 8) | TH_2316 | - | 5e-10 | 30.67% (225) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0230 | - | 8e-08 | 24.91% (277) | hypothetical protein MUL_0230 |
| M. vanbaalenii PYR-1 | Mvan_6043 | - | 2e-22 | 32.82% (323) | hypothetical protein Mvan_6043 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0912|M.bovis_AF2122/97 -------------------MDYAKRIGQVGALAVVLGVGAAVTT-HAIGS
Rv0888|M.tuberculosis_H37Rv -------------------MDYAKRIGQVGALAVVLGVGAAVTT-HAIGS
MMAR_4644|M.marinum_M -------------------MGYASHAVRVGALVFVLVVGAAVATDQAIGH
MAV_1185|M.avium_104 --------------------------------------------------
MUL_0230|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 -------------------------MGRIGALAVALGIGAALTTG-TGVA
TH_2316|M.thermoresistible__bu -------------------------VGRVGALAVALGVGVAVATGGTAVA
Mflv_0780|M.gilvum_PYR-GCK --MAGAHRAKPLPTRGRSQSRRLLPYTWLGAGAVALGVGAALTGGTAPAH
Mvan_6043|M.vanbaalenii_PYR-1 MPQERELPLPQRRRRGARRAEDLPVRSWLKLGAASAGLGAALLGFALAGP
Mb0912|M.bovis_AF2122/97 AAPTDPSSSSTDSPVDACSPLGG---SASSLA------------------
Rv0888|M.tuberculosis_H37Rv AAPTDPSSSSTDSPVDACSPLGG---SASSLA------------------
MMAR_4644|M.marinum_M AAPTDSDASNGESWADVPAPPGGENIAASLLPG-----------------
MAV_1185|M.avium_104 --------------------------------------------------
MUL_0230|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 WADDSGASGGSTSSSSSASDS------SSSSAGGSSGSASG---------
TH_2316|M.thermoresistible__bu WAD-SGSTASSSSSQDSSSKSNPAQTRSSAKSGIGSGAGSGPGSRLDRPA
Mflv_0780|M.gilvum_PYR-GCK ADSGGESSQTHGESRQSRASRDTAERPSQTRSADDGAGSNSDDRSTDDER
Mvan_6043|M.vanbaalenii_PYR-1 SPMAEADTGTETSVSTGPEKATAEQSGSGEADAPDDAEDEAN--------
Mb0912|M.bovis_AF2122/97 --------------------------------------------------
Rv0888|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4644|M.marinum_M ---------------------------------------------AAQPG
MAV_1185|M.avium_104 ----------------------------------------------MNPG
MUL_0230|M.ulcerans_Agy99 ----------------------------------------------MS-D
MSMEG_4365|M.smegmatis_MC2_155 --------GADAAKPRKPGVTKP------------RIQPTGKIAESLKKT
TH_2316|M.thermoresistible__bu SRAPLGGSGADSSATEQTGAVSSGTSAWRSAGMPAGTSLTGRLADSVRDA
Mflv_0780|M.gilvum_PYR-GCK PLATGNKTRLSKTTRESSSGEARFADAVSEEAHTGAETPASTETSGLEVR
Mvan_6043|M.vanbaalenii_PYR-1 ---------------------------------------EDSDELDDADA
Mb0912|M.bovis_AF2122/97 ---------------AIPGASVPQVGVRQVDPGSIPDDLLNALIDFLAAV
Rv0888|M.tuberculosis_H37Rv ---------------AIPGASVPQVGVRQVDPGSIPDDLLNALIDFLAAV
MMAR_4644|M.marinum_M AVPGAALPGTALPGTALPGAASPGMGTPQLDPSNIPDDLLNAVINFLASI
MAV_1185|M.avium_104 ANSPRPYRRLRHLATRADKHSKGGTSVSDANGPTARFGIDNTAVVKVPVH
MUL_0230|M.ulcerans_Agy99 VNGQQ---------TGWNDRDGGDVQVFELG--TAP---DFPTSVEIAVA
MSMEG_4365|M.smegmatis_MC2_155 AKAVEGAVADAHKRAQQSAAQQAEKRAERRAKLRAQIRPGKDTDKQAPEK
TH_2316|M.thermoresistible__bu VKPWRPATTRSTDPTPVETPAEANPEAQPDAHAGSPGESTAPETQPAPSS
Mflv_0780|M.gilvum_PYR-GCK PGDAATDNPDDTAVRAAALAPAARTPSTQDDGTPDAQRTAHVHSGALPAA
Mvan_6043|M.vanbaalenii_PYR-1 TDDPDDPDDADATDDVDDVDEADDAVGEATDGDRDDEGGEAAAGGADPAA
.
Mb0912|M.bovis_AF2122/97 RN------------------------------------------------
Rv0888|M.tuberculosis_H37Rv RN------------------------------------------------
MMAR_4644|M.marinum_M RN------------------------------------------------
MAV_1185|M.avium_104 HG------------------------------------------------
MUL_0230|M.ulcerans_Agy99 NG------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 Q-------------------------AGNEAKNAAQPATKTQRQLP----
TH_2316|M.thermoresistible__bu RGIRSMTA----------------VLPDTESTNTESTNTESTVQHDDSER
Mflv_0780|M.gilvum_PYR-GCK EGNRASAAGSSLGALFRNQTPTVSWTQASPTGSSVVTGVLTASDPDGDAL
Mvan_6043|M.vanbaalenii_PYR-1 DG------------------------------------------------
Mb0912|M.bovis_AF2122/97 ---------------------GLVPIIENRTPVANPQQVSVPEGG-----
Rv0888|M.tuberculosis_H37Rv ---------------------GLVPIIENRTPVANPQQVSVPEGG-----
MMAR_4644|M.marinum_M ---------------------GVVPILENRTPVATPQQLEIDVNG-----
MAV_1185|M.avium_104 ------------------------PISDIDVSPDGRRLLVTNYGR-----
MUL_0230|M.ulcerans_Agy99 ------------------------PIGDIGISPDGSRLMVTNYGD-----
MSMEG_4365|M.smegmatis_MC2_155 ----AAPAVKVPDVITSVRARSDEVAREITTGVTGIRTAIENAAPRIALP
TH_2316|M.thermoresistible__bu RSSLRSTAASVPELSTPVRLADATAVRALAERLTERRSVTLESVR--AFT
Mflv_0780|M.gilvum_PYR-GCK SYKVTRNPAYGAVTVYADGSVAYTPHQNVATSIDSFRVTVSDAASGFHVH
Mvan_6043|M.vanbaalenii_PYR-1 -----------------------DVVTDLPALVESPPPTENSGAT-----
Mb0912|M.bovis_AF2122/97 -------------------------------------------------T
Rv0888|M.tuberculosis_H37Rv -------------------------------------------------T
MMAR_4644|M.marinum_M -------------------------------------------------S
MAV_1185|M.avium_104 --------------------------------------------------
MUL_0230|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_4365|M.smegmatis_MC2_155 DLRPTTNL---------------------------------------ARP
TH_2316|M.thermoresistible__bu DVAATTALTSA--------------------------------ASVAAAT
Mflv_0780|M.gilvum_PYR-GCK GLSGLINLFSFGLLGKSGHEHTITAEVRFAATAVPQPTVLDRIVLPLNAG
Mvan_6043|M.vanbaalenii_PYR-1 -------------------------------------------------T
Mb0912|M.bovis_AF2122/97 VGPVRFDACDPDG--------------------NRMTFAVRERGAPGGPQ
Rv0888|M.tuberculosis_H37Rv VGPVRFDACDPDG--------------------NRMTFAVRERGAPGGPQ
MMAR_4644|M.marinum_M TGLVPFSAYDPDG--------------------NRMTFKVNPRGTPGGPQ
MAV_1185|M.avium_104 -DTVSVIDTHTCR-----------------VASTIAGLSEPFAVAMSAAD
MUL_0230|M.ulcerans_Agy99 -NSVSVIDTDTCR-----------------VVETITGLDEAFAVTAGGPA
MSMEG_4365|M.smegmatis_MC2_155 VTPVALKAAQPVQ---VSPPRPTITGLVRGLLTAAAGFSPRAATAPVAPA
TH_2316|M.thermoresistible__bu VEPVALITAAPEATPAVDAEQPAAPRLVTGLL-AAVGLAPFTANSPLAPT
Mflv_0780|M.gilvum_PYR-GCK QSPRTVAVSPDGRLVFVGSGRWNSPCCGFDNPDGALSVIDAASGAVLSTV
Mvan_6043|M.vanbaalenii_PYR-1 ESAASAQPATPKR---------PWRELVAGFIDDWTADTEAWIDSLDASD
: ..
Mb0912|M.bovis_AF2122/97 HG-----IVTVDQRTASFIYTADPGFVGTDTFSVNVSDDTSLHVHGLAGY
Rv0888|M.tuberculosis_H37Rv HG-----IVTVDQRTASFIYTADPGFVGTDTFSVNVSDDTSLHVHGLAGY
MMAR_4644|M.marinum_M HG-----MVNIDESRAAFVYTPEPNFVGIDTFTVAVSDDTSLHVHGLAGY
MAV_1185|M.avium_104 PN---------------YAYVSTATAAYDAIEVIDVVTNWRIATHRLAHS
MUL_0230|M.ulcerans_Agy99 AN---------------RAYVSTVSTAYDAIAVIDVATNAVVSTHPLALS
MSMEG_4365|M.smegmatis_MC2_155 PSPAAWALLAFARREFERAFTPGAATALAAPAITTSAVTSAAVVNPLPGR
TH_2316|M.thermoresistible__bu QSTPLLALLGWARRETDRAYTQQARSLVTA---RPAVVSTAAVVDPLPAD
Mflv_0780|M.gilvum_PYR-GCK KTGLTPISFAVGSTGLLYAVGVDGGTGYTNYFLSAMTPNDYLTKSTFTDT
Mvan_6043|M.vanbaalenii_PYR-1 QTKERLEATFWAVRRTFFNQAPTVDPVQITGVITGPVTGTLGAVDPDGDR
Mb0912|M.bovis_AF2122/97 LG-----------------PFHGHDDVATVTVFVGN----------TPTD
Rv0888|M.tuberculosis_H37Rv LG-----------------PFHGHDDVATVTVFVGN----------TPTD
MMAR_4644|M.marinum_M LG-----------------PFHGHDDVATVTIFVGN----------IPTG
MAV_1185|M.avium_104 LS-------------------DLAVSPDGRYLYAS-------------RN
MUL_0230|M.ulcerans_Agy99 VS-------------------DLAASSDGKYVYAS-------------RN
MSMEG_4365|M.smegmatis_MC2_155 LD--SVPVGWVTGVSNPSGNWPQTNNTAGFDIWGTDLG---IMWDGGEWN
TH_2316|M.thermoresistible__bu LIGETERIGRITGPG-------ITDGPNRLNIYGTDLG---IMWDMGEWR
Mflv_0780|M.gilvum_PYR-GCK RAIP----------DLYMQPISLAVTPDGNSVVIGGRNGQVAVVDPMTTR
Mvan_6043|M.vanbaalenii_PYR-1 VVYRLVRAP---------GQGSVKVNSDGTFTYTPGSD-------FDGVD
Mb0912|M.bovis_AF2122/97 TISGDFSMLTYNIAG-----------------------------------
Rv0888|M.tuberculosis_H37Rv TISGDFSMLTYNIAG-----------------------------------
MMAR_4644|M.marinum_M TISGDFSMLTYNIAG-----------------------------------
MAV_1185|M.avium_104 AVRGADVAVLDTTTG-----------------------------------
MUL_0230|M.ulcerans_Agy99 GARGADIAVLDTATG-----------------------------------
MSMEG_4365|M.smegmatis_MC2_155 GQRFVHTAFGDTFSGPNMSGWWRSNVLLISTDNVLSDGLG--LVQTGTAY
TH_2316|M.thermoresistible__bu GQRAVHVVFGDTFSGPNMSGDWISNAIMISTSRTLYENPAGFLEQTGPAF
Mflv_0780|M.gilvum_PYR-GCK VIRTFDIGFGNHAEGVYVTPDGKYAWISGGYYQPLFRLDLSTGAIGQVQI
Mvan_6043|M.vanbaalenii_PYR-1 TFRVLAVDVGMHVNVLDWFRPWG---------------------------
.
Mb0912|M.bovis_AF2122/97 ------------------------------LPFPLSSAILPRFFY-----
Rv0888|M.tuberculosis_H37Rv ------------------------------LPFPLSSAILPRFFY-----
MMAR_4644|M.marinum_M ------------------------------LPFPLSSAMLPRFLY-----
MAV_1185|M.avium_104 --------------------------ELEVIELAAAAGTTTACVR-----
MUL_0230|M.ulcerans_Agy99 --------------------------RVDVVDIATKPGLTTECVR-----
MSMEG_4365|M.smegmatis_MC2_155 QFIPA---------SGRNLFGLFGGSEVTVIPTAGVQIDGTQYVN-----
TH_2316|M.thermoresistible__bu QFIP-----------GGQLLSWFFPAEVTVIPTAGVHANGTQYVN-----
Mflv_0780|M.gilvum_PYR-GCK DYGAGGVLSPDGSKFYTVVDRHDYGQRIEIVDTATGAISSYDYRSESVHR
Mvan_6043|M.vanbaalenii_PYR-1 ---------------------------TRATPLINQGAVTFDFRYETGAG
Mb0912|M.bovis_AF2122/97 ------TKEIGKRLNAYYVANVQEDFAYHQFLIKKSKMPSQTPPEPPTLL
Rv0888|M.tuberculosis_H37Rv ------TKEIGKRLNAYYVANVQEDFAYHQFLIKKSKMPSQTPPEPPTLL
MMAR_4644|M.marinum_M ------TKEIGKRLNNFSVSNVQEDFSYHEFLIKKSRMPSQTPPSPPTLL
MAV_1185|M.avium_104 ------VSADGRRLFVGVNGPNGGGLAIIET------------RTRTDGR
MUL_0230|M.ulcerans_Agy99 ------VSADGNRVDVGANGPSGGQLIVVATHAESELNPVRGGRARSRGR
MSMEG_4365|M.smegmatis_MC2_155 ------YMSVRSWDTPGRWTTNFSAISVYDPSTDTWVLAPSTIRSAGWFR
TH_2316|M.thermoresistible__bu ------YMSVRQWGVPGSWTTNFSAISRYDQATDTWVTVPSSVRSAGWLR
Mflv_0780|M.gilvum_PYR-GCK NLIDLAVSPDGKHLYARIAASQTSNRLDVIDAATGVVVAQTQVGSEAALL
Mvan_6043|M.vanbaalenii_PYR-1 -----HWSAERKAALLGAANELVSYLRVNAPVVISYRVDGFDDPTSALLA
Mb0912|M.bovis_AF2122/97 WPIGVPFSDGLNTLSEFKVQRLDRQT---------------------WYE
Rv0888|M.tuberculosis_H37Rv WPIGVPFSDGLNTLSEFKVQRLDRQT---------------------WYE
MMAR_4644|M.marinum_M WPIGVPFSDGLNSLAVFKVKRLDRQT---------------------WFQ
MAV_1185|M.avium_104 RVGGRS-----------RLVGTVELG----------------------LP
MUL_0230|M.ulcerans_Agy99 NGQGVAGSVVQQFPRDLRIIDTIEIG----------------------SS
MSMEG_4365|M.smegmatis_MC2_155 SSTLYVPGSQNFQQAAYVLQPEDQVGEDGIRYLYAFGTPSGRAGSAYLSR
TH_2316|M.thermoresistible__bu SSTPYRAGDQNFQQMAYVLQPVDQVGEDGVRYVYAFGTPSGRQGSVHLSR
Mflv_0780|M.gilvum_PYR-GCK GLPGMAISPEGNRIYLATGDSITVVRANSTDRPLLPRAFPSASASALLLN
Mvan_6043|M.vanbaalenii_PYR-1 QAGSSYTSAEPGFWPTLVQNKLISGGD--------------------ANG
Mb0912|M.bovis_AF2122/97 CTSDNCLTLKGFTYSQMR------------LPGGDTVDVYNLHTNTGGGP
Rv0888|M.tuberculosis_H37Rv CTSDNCLTLKGFTYSQMR------------LPGGDTVDVYNLHTNTGGGP
MMAR_4644|M.marinum_M CDADNCLTPKGFSYSQLQ------------LPGGNTMDLYNLHANTGGGV
MAV_1185|M.avium_104 VRDVALGNDGNTAYVAS----------CGPVVGSVLDVVDTRAATIVSTH
MUL_0230|M.ulcerans_Agy99 VRDVAISPDGTTAYVAS----------CGADFGAVIDVVDTRTHKITGTR
MSMEG_4365|M.smegmatis_MC2_155 VPEDAVTDLSKYEYWDGNRWVTGNAAVAAPVIGDSTRSTGLFGFIVDWAN
TH_2316|M.thermoresistible__bu VPEEHITDVSKYEYWDGNAWVS-SAAHAAPIIGESTRSAGLFGFIIDWAN
Mflv_0780|M.gilvum_PYR-GCK LPRQTDTIYAEKVMRDGKTR----MIVYMTGIDAGLNESTLASIISNSGL
Mvan_6043|M.vanbaalenii_PYR-1 SAADGLIQVNFGKKWSLDDTVESDEFDFMMALMHEMLHSFGILFAVRPGE
Mb0912|M.bovis_AF2122/97 TTNANLAQVANYIQQNSAGRAVIVTGDFNARYSDDQSALLQFAQVNGLTD
Rv0888|M.tuberculosis_H37Rv TTNANLAQVANYIQQNSAGRAVIVTGDFNARYSDDQSALLQFAQVNGLTD
MMAR_4644|M.marinum_M LTNANLRQLANYILQNSAGRAVIVTGDFNALYSEDQAGLLDFAGSTGLTD
MAV_1185|M.avium_104 KINEITGPLTRMTLSRDGERAYLISDDRVTVLGTRTQDVLGEVRVTKHPS
MUL_0230|M.ulcerans_Agy99 KIEEIGGLITRLSIGGNGERAYLVSEDRVTVLSARSQDVIGTVRVN-QPS
MSMEG_4365|M.smegmatis_MC2_155 DPNVLGGYLGGLVGAKTGGNVSELSVQYNEYLG-KYVVLYGDG-NNNIQM
TH_2316|M.thermoresistible__bu DPNVFGGYLGGLFGAKTGGNVGEMSVQYNEYLG-KYVILYGNGRTNNIEM
Mflv_0780|M.gilvum_PYR-GCK LHADVEAYIDNAYTAWGGARVIEEIQLVGHSNGGQQMQAYADWGTYADKV
Mvan_6043|M.vanbaalenii_PYR-1 TTRRYWSTYASLVVDENGSSPFNSDFSWNTAYDPNLTRKDGGLFFGGTEA
. .
Mb0912|M.bovis_AF2122/97 AWVQVE-------HGPTTPPFAPTCMVGNECELLDKIFYRS---GQGVTL
Rv0888|M.tuberculosis_H37Rv AWVQVE-------HGPTTPPFAPTCMVGNECELLDKIFYRS---GQGVTL
MMAR_4644|M.marinum_M AWVEVE-------HGPTTPPFAPDCMVGNECELLDKIFYRS---GTGVTL
MAV_1185|M.avium_104 ALVESP-------DGNY------LYVADHAG--VLSVARLP---SGTAPA
MUL_0230|M.ulcerans_Agy99 AAIESP-------GGDR------LYVADYSGTVTMTLVDLG---VSAIDN
MSMEG_4365|M.smegmatis_MC2_155 RIADRP-------EGPWSDPIELASSADYPG--LYAPMIHP---WSGTGM
TH_2316|M.thermoresistible__bu RIADSP-------EGPWSDPIILATPQQYPG--LYAPMIHP---WSGTGY
Mflv_0780|M.gilvum_PYR-GCK TSVVLFGAPIIKTEDEFTSDALAFINRGDPVPTIFSHWENPGVRWNSMNK
Mvan_6043|M.vanbaalenii_PYR-1 VEAYDG--------ALIPLFTSDPWLGTSVGAHLDDMVFIG--VDRKMMN
Mb0912|M.bovis_AF2122/97 QAVSYGNEAPKFFNSKGEPLSDHSPAVVGFHYVADNVAVR----------
Rv0888|M.tuberculosis_H37Rv QAVSYGNEAPKFFNSKGEPLSDHSPAVVGFHYVADNVAVR----------
MMAR_4644|M.marinum_M QATAYSNEAPNFNNSNGQPLSDHSPPAVAFHYETTNVSYPN---------
MAV_1185|M.avium_104 APCSGADEDDDATTGWLPELAPWEPVLA----------------------
MUL_0230|M.ulcerans_Agy99 ALCQGLSAD-----WVMPELLQFEAALA----------------------
MSMEG_4365|M.smegmatis_MC2_155 LEDDNGDPDVNNLYWNMSLWGDYNVVLMQTDLSGLKTVAV----------
TH_2316|M.thermoresistible__bu LLDENGDPDPESLYWNMSMWGPYNVYLMQTDLSSLVEV------------
Mflv_0780|M.gilvum_PYR-GCK DINFYGPESSDALNFERHHASSYYAAAVDFDAKAGLSSAPATQRRLRDTL
Mvan_6043|M.vanbaalenii_PYR-1 ASTTPGPGVRVLSAIELGILKDLGYQVVARTPVEDLA-------------
Mb0912|M.bovis_AF2122/97 -------------------------
Rv0888|M.tuberculosis_H37Rv -------------------------
MMAR_4644|M.marinum_M -------------------------
MAV_1185|M.avium_104 -------------------------
MUL_0230|M.ulcerans_Agy99 -------------------------
MSMEG_4365|M.smegmatis_MC2_155 -------------------------
TH_2316|M.thermoresistible__bu -------------------------
Mflv_0780|M.gilvum_PYR-GCK LRFGGYPVDAMPTAHIAVFTSVTRV
Mvan_6043|M.vanbaalenii_PYR-1 -------------------------