For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKIGIIGAGHIGGTLTRRLRELGHEVNVANSRSPETLADLERETGATAVWAKDAAADADLVIVSIPQKNV PDLADGIVDARKPDAPVIETNNYYPQQRDGEIAAIEDGQPESAWVAEQIGAPVYKVFNGIFWKHLLERGI PAGSPGRIALPVAGEDGPNRELVHIVVDQLGFDPVDAGPVSESWRQQPGTPVYGKDFGVEKTRAALAEAA PERTAEWSARTA
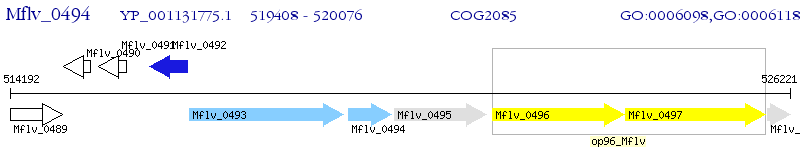
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0494 | - | - | 100% (222) | NADP oxidoreductase, coenzyme F420-dependent |
| M. gilvum PYR-GCK | Mflv_5012 | - | 4e-05 | 30.77% (156) | 3-hydroxyisobutyrate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5884 | - | 2e-08 | 31.62% (136) | 3-hydroxyisobutyrate dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_0955 | proC | 4e-05 | 27.45% (153) | PROBABLE PYRROLINE-5-CARBOXYLATE REDUCTASE PROC (P5CR) (P5C |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1360 | - | 2e-05 | 30.13% (156) | 3-hydroxyisobutyrate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5884|M.smegmatis_MC2_155 -MRIGFIGLGNMGAAMAANLLAAGHDVV--AYN-RSPQKVAALAEQGATA
Mvan_1360|M.vanbaalenii_PYR-1 MTTIAFLGLGNMGGPMAANLVAAGQTVH--AFD-LVTELKEAAKAKGAKV
Mflv_0494|M.gilvum_PYR-GCK -MKIGIIGAGHIGGTLTRRLRELGHEVN--VANSRSPETLADLERETGAT
TH_0955|M.thermoresistible__bu VARIAIIGGGNIGEALLSGLLRAGRQVKDLVVAEKDPNRAKYLAEKYGVL
*.::* *::* .: * *: * . .: : .
MSMEG_5884|M.smegmatis_MC2_155 AES-VRDAAQADVVVTMLADDNAVETVAFGGADPAEGLVAAMPADAIHIS
Mvan_1360|M.vanbaalenii_PYR-1 FDSGAAAVADADVVITSLPNGNIVKAVYD-------EVVPAAKAGALLID
Mflv_0494|M.gilvum_PYR-GCK AVWAKDAAADADLVIVSIPQKNVPDLADG--------IVDARKPDAPVIE
TH_0955|M.thermoresistible__bu VTSVGDAAESATFIVIAVKPGDVEKVIDD-------IAEAAARADSGTDE
. .* .:: : : . * ..: .
MSMEG_5884|M.smegmatis_MC2_155 SSTISVDLAKRL---AAAHAEARNQFVSAPVFGRPDAAAAAKLFVVASGA
Mvan_1360|M.vanbaalenii_PYR-1 TSTISVDDARAI---HDQATEKGLAQIDAPVSGGIKGATAGTLAFMVGGE
Mflv_0494|M.gilvum_PYR-GCK TNNYYPQQRDGE---IAAIEDG------QPESAWVAEQIGAPVYKVFNG-
TH_0955|M.thermoresistible__bu QVFVSVAAGVGTGYYEAKLPAGAPVVRVMPNAPMVVGGGVSALAPGRFAT
* . : .
MSMEG_5884|M.smegmatis_MC2_155 PAAVEAATPVFDAIGQRTFVVGDEPSAASLVKISGNFLIASVIESLGEAM
Mvan_1360|M.vanbaalenii_PYR-1 DEAFERAKPVLEPMAGKIIHCG-ASGAGQAAKLCNNMVLAVQQIAIGEAF
Mflv_0494|M.gilvum_PYR-GCK --------IFWKHLLERGIPAG----------------------SPGRIA
TH_0955|M.thermoresistible__bu PEQLKEVAGIFDAVGGVLTVTEAQMDAVTAVSGSGPAYFFLMAEAMVDAG
. . : :
MSMEG_5884|M.smegmatis_MC2_155 ALVGKAGVDKQQYLELLT-------STLFDAPVYRTYGGLLVDEQFTPPG
Mvan_1360|M.vanbaalenii_PYR-1 VLAEKLGLPAQSLFDVITGATGNCWAVHTNCPVPGPVPTSPANNDFKP-G
Mflv_0494|M.gilvum_PYR-GCK LPVAGEDGPNRELVHIVV-----------DQLGFDPVDAGPVSESWRQ-Q
TH_0955|M.thermoresistible__bu VAVGLSRAVATGLVAQTMAG---SAAMLLDRLDDAAKSAAGAAMDTSPAE
. . : . . .
MSMEG_5884|M.smegmatis_MC2_155 FAAPLGLKDVKLALSAGEELRVPLPVGSLLRDRFLTLLATGGGELDWSAL
Mvan_1360|M.vanbaalenii_PYR-1 FATALMNKDLGLAMAAVSSTGSSAPLGTHAAEIYAEFNKEHADKDFSAII
Mflv_0494|M.gilvum_PYR-GCK PGTPVYGKDFGVEKTR-------AALAEAAPERTAEWSARTA--------
TH_0955|M.thermoresistible__bu LRAMVTSPGGTTAAGLRELERGGLRAAVANAVEAAKMRSEQLGITSE---
: : . .
MSMEG_5884|M.smegmatis_MC2_155 GALPAWEAGAPHPA
Mvan_1360|M.vanbaalenii_PYR-1 EALRG---------
Mflv_0494|M.gilvum_PYR-GCK --------------
TH_0955|M.thermoresistible__bu --------------