For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAEDGPRGLSHLKVDRRAEVPDGTTSFYYRTRTALLHGVADQLVRYDAEAFTDAFETAPLDDGDVIAAQL ARQILSIRDEPQLSRTRARLELTLLSRNDPVISANFQQIGESMRALVERLVVAVQAGSRPLERGLLDEQV SVVLSYLGGLILAFANGSPEPLSGEDISRQFRAVIRGVAAERADRRREPAQSSSF*
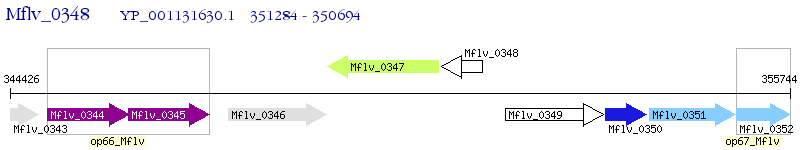
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0348 | - | - | 100% (196) | hypothetical protein Mflv_0348 |
| M. gilvum PYR-GCK | Mflv_3484 | - | 1e-20 | 36.11% (180) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4710c | - | 4e-13 | 32.28% (189) | TetR family transcriptional regulator |
| M. marinum M | MMAR_5226 | - | 6e-07 | 35.64% (101) | DeoR family transcriptional regulator |
| M. avium 104 | MAV_5252 | - | 4e-27 | 39.25% (186) | hypothetical protein MAV_5252 |
| M. smegmatis MC2 155 | MSMEG_0656 | - | 6e-25 | 39.01% (182) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4301 | - | 2e-07 | 29.05% (148) | DeoR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0389 | - | 2e-66 | 69.57% (184) | hypothetical protein Mvan_0389 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0348|M.gilvum_PYR-GCK -------------------------MLAEDGPRGLSHLKVDRRAEVPDGT
Mvan_0389|M.vanbaalenii_PYR-1 MTRVDSRRRPNAAQRRHQLCDAANTLLAEDGPRGLSHLKVDRRAGVPDGT
MAV_5252|M.avium_104 -------------------------MLGGNGLHGVSHPKVDDRAGVPAGT
MSMEG_0656|M.smegmatis_MC2_155 -----MGRKQNPEQRRRELCDAAIRLLADEGIKGLTHLKVDRTAGVPDGT
MMAR_5226|M.marinum_M --------MPPNPQRRAQILDAAIDILADVGVGGLTHRQVDDRAGLPAGT
MUL_4301|M.ulcerans_Agy99 --------MPPNPQRRAQILDAAIDILADVGVGGLTHRQVDDRAGLPAGT
MAB_4710c|M.abscessus_ATCC_199 ----------MADDRRTQLADAGLAVLAAGGARGLTHRAVDRSAELPEGS
:*. * *::* ** * :* *:
Mflv_0348|M.gilvum_PYR-GCK TSFYYRTRTALLHGVADQLVRYDAEAFTDAFETAPLDDGDVIAA-QLARQ
Mvan_0389|M.vanbaalenii_PYR-1 TSFYYRTRVALLQGVADHLARYDAEAFAQAFDDVPDGAGDDLLP-MLAEQ
MAV_5252|M.avium_104 TSFYFRTRKALVHAMAGRLAELDVADFSMMAELAEDHATEFAGTAGLARI
MSMEG_0656|M.smegmatis_MC2_155 TSFYFRTSSALLHAVAGRVAELDLQDLTAATEPPAPGENPAAG---LAAL
MMAR_5226|M.marinum_M TSNYFRTRQALLEATASRTVDLHWQRVQVLRSAVGSLSRDGVKA--LMTK
MUL_4301|M.ulcerans_Agy99 TSNYFRTRQALLEATASRTVDLHWQRVQVLRSAVGSLSRDGVKA--LMTK
MAB_4710c|M.abscessus_ATCC_199 TSYYLRTRVALLQACATRLGERTMTAVVPLAEPVHLDISALTQ---LAVR
** * ** **:.. * : . . *
Mflv_0348|M.gilvum_PYR-GCK ILSIRDEPQLSRTRARLELTLLSRNDPVISANFQQIGESMRALVERLVVA
Mvan_0389|M.vanbaalenii_PYR-1 VLLVRDEPHLSRTRARLELTMLAKGDAGIAAGFQQVSESYRALTERLVTA
MAV_5252|M.avium_104 VMYVNSEPWLTRAKARYELALLAGRDPELAAALSESADRLYALARNVVTQ
MSMEG_0656|M.smegmatis_MC2_155 VMRAADGPGFVRSRARLELAMPSSRDPGLAQVFALHQQRFVDMHRDVVQR
MMAR_5226|M.marinum_M MLTDPDEQFRRNTLARFELFMEGTRRPELQPFLVELQR--AAVKSATLIF
MUL_4301|M.ulcerans_Agy99 MLTDPDEQFRRNTLARFELFMEGTRRPELQPFLVELQR--AAVKSATLIF
MAB_4710c|M.abscessus_ATCC_199 AVRAWIADGGVLVLARHELLLESAQHPQMRSVLDAATDHIRGLLERRLIA
: ** ** : . . : : : :
Mflv_0348|M.gilvum_PYR-GCK VQAG-SRPLERGLLDEQVSVVLSYLGGLILAFANGSPEPLSGEDISRQFR
Mvan_0389|M.vanbaalenii_PYR-1 MQNGLTAPLDRSLLDEQVSVVLTYLSGLVFAFANGSTEPMTSRDIQRQIR
MAV_5252|M.avium_104 WHPAGSA-PDPALVDDQATATLAFINGIMLTFVAGQPAVDDAGQLDRLIQ
MSMEG_0656|M.smegmatis_MC2_155 LSPD----ADPAEVAERTYVLLTFISGLLLALSRGDRTIGSAEQLEAIIC
MMAR_5226|M.marinum_M EAAG------FAPTPEQMDQLSRLLNGYVFSHLTLPGDDDPAALVDRLLP
MUL_4301|M.ulcerans_Agy99 EAAG------FAPTPEQMDQLIRLLNGYVFSHLTLPGDDDPAALVDRLLP
MAB_4710c|M.abscessus_ATCC_199 LGIS--------DAAERTGDLIACLDGIALAYAVRG--VAAEDALRRSVT
:: :.* :: : .
Mflv_0348|M.gilvum_PYR-GCK AVIRGVAAERADRRREPAQSSSF
Mvan_0389|M.vanbaalenii_PYR-1 AVVIGVAAERLS------QTS--
MAV_5252|M.avium_104 GVIAGVAHVRGA-----------
MSMEG_0656|M.smegmatis_MC2_155 SLCRGGPQPEQG-----------
MMAR_5226|M.marinum_M AYFDA------------------
MUL_4301|M.ulcerans_Agy99 AYFDA------------------
MAB_4710c|M.abscessus_ATCC_199 RVVSGLLAVRD------------
.