For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSAAMRGPTAMTENMIVTATVDAPVSAVFSVLADPAQHAAIDGTGWVREGLDGEFLSRAGQLFRMAMYHD NVPNGHYEMVNKVIAFERDREIGWEPGQAGPDGVVGYGGWTWHYTLEPIGPAQTRVTLTYDWSGVPAELR EHIEFPPFPDTHLKNSLDNLATLVVG*
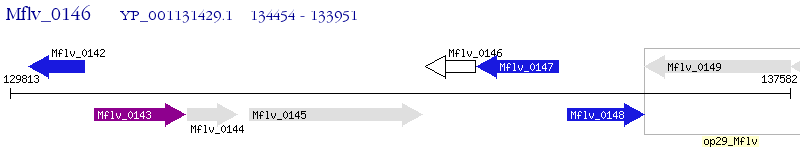
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0146 | - | - | 100% (167) | hypothetical protein Mflv_0146 |
| M. gilvum PYR-GCK | Mflv_3232 | - | 1e-21 | 35.57% (149) | hypothetical protein Mflv_3232 |
| M. gilvum PYR-GCK | Mflv_2610 | - | 5e-12 | 29.63% (135) | hypothetical protein Mflv_2610 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0091 | - | 6e-08 | 27.35% (117) | hypothetical protein Mb0091 |
| M. tuberculosis H37Rv | Rv0088 | - | 6e-08 | 27.35% (117) | hypothetical protein Rv0088 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2457c | - | 1e-11 | 28.85% (156) | hypothetical protein MAB_2457c |
| M. marinum M | MMAR_0253 | - | 6e-12 | 31.13% (151) | hypothetical protein MMAR_0253 |
| M. avium 104 | MAV_5230 | - | 2e-12 | 30.00% (150) | hypothetical protein MAV_5230 |
| M. smegmatis MC2 155 | MSMEG_3417 | - | 1e-45 | 55.92% (152) | hypothetical protein MSMEG_3417 |
| M. thermoresistible (build 8) | TH_4053 | - | 2e-42 | 52.15% (163) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4861 | - | 4e-12 | 31.13% (151) | hypothetical protein MUL_4861 |
| M. vanbaalenii PYR-1 | Mvan_0758 | - | 6e-64 | 69.23% (169) | hypothetical protein Mvan_0758 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0146|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0758|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3417|M.smegmatis_MC2_155 --------------------------------------------------
TH_4053|M.thermoresistible__bu --------------------------------------------------
MMAR_0253|M.marinum_M --------------------------------------------------
MUL_4861|M.ulcerans_Agy99 --------------------------------------------------
MAB_2457c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5230|M.avium_104 --------------------------------------------------
Mb0091|M.bovis_AF2122/97 MSVYKHAPSRVRLRQTRSTVVKGRSGSLSWRRVRTGDLGLAVWGGREEYR
Rv0088|M.tuberculosis_H37Rv MSVYKHAPSRVRLRQTRSTVVKGRSGSLSWRRVRTGDLGLAVWGGREEYR
Mflv_0146|M.gilvum_PYR-GCK ------------MSSAAMRGPTAMTENMIVTATVDAPVSAVFSVLADPAQ
Mvan_0758|M.vanbaalenii_PYR-1 ------------MSPACDRGLTRMTETLIVDTIVDAPASTVFAVLADPAT
MSMEG_3417|M.smegmatis_MC2_155 ------------MSNDIMTAER----------IVDAPAATVFGVLADPTA
TH_4053|M.thermoresistible__bu ------------MS-DIQTTGTSL----AVHTTVNAPARTVFDVLADPTT
MMAR_0253|M.marinum_M ------------MVDTDAPLPDRSERVVSATREIAAGAAQIFELIADPSR
MUL_4861|M.ulcerans_Agy99 ------------MVDTDAPLPDRSERVVSATREIAAGAAQIFELIADPSR
MAB_2457c|M.abscessus_ATCC_199 ------------MTD--------DVRVVSASREVQASAAAIFELIADPAR
MAV_5230|M.avium_104 ------------MSDT------EAERIVSATRVIAANAARIFELIADPAR
Mb0091|M.bovis_AF2122/97 AVKPGTPGIQPKGDMMTVTVVDAGPGRVSRSVEVAAPAAELFAIVADPRR
Rv0088|M.tuberculosis_H37Rv AVKPGTPGIQPKGDMMTVTVVDAGPGRVSRSVEVAAPAAELFAIVADPRR
: * . :* ::***
Mflv_0146|M.gilvum_PYR-GCK HAAIDGTGWVREGLDGEFLSRAGQLFRMAMYHDN--VPNGHYEMVNKVIA
Mvan_0758|M.vanbaalenii_PYR-1 HAAIDGTGWVRESLDGEPITRSGQMFRMAMYFANPAFADGHYEVANKVIA
MSMEG_3417|M.smegmatis_MC2_155 HHAIDGTGWVRESLDSAKLTTVGQIFRMAMYHRN--FGGMHYTVANRVEV
TH_4053|M.thermoresistible__bu HARIDGTGWVREPVDGARLTGAGQIFRMGMYHEN--HPHRHYEIANRVEV
MMAR_0253|M.marinum_M QPDWDGNDNLAHCAAGQRVHRVGDVFRMTLTLGS--------VRENHIVE
MUL_4861|M.ulcerans_Agy99 QPDWDGNDNLAHCAAGQRVHRVGDVFRMTLTLGS--------VRENHIVE
MAB_2457c|M.abscessus_ATCC_199 QREWDGNENLAEAAAGQRAHAVGDVFVMTLTTGA--------DRENHIVE
MAV_5230|M.avium_104 QPGWDGNDNLASAAPGQRVHRVGDVFKMTLTNGA--------VRENHVTE
Mb0091|M.bovis_AF2122/97 HRELDGSGTVRGNIKVPAKLVVGSKFSTKMKLFG-----LPYRITSRVTA
Rv0088|M.tuberculosis_H37Rv HRELDGSGTVRGNIKVPAKLVVGSKFSTKMKLFG-----LPYRITSRVTA
: **. : *. * : .::
Mflv_0146|M.gilvum_PYR-GCK FERDREIGWEPGQAG---------PDGVVGYGGWTWHYTLEPIG----PA
Mvan_0758|M.vanbaalenii_PYR-1 FEPDRAIGWEPGQAR---------EDGTVEHGGWTWRYDLEAVG----PG
MSMEG_3417|M.smegmatis_MC2_155 FDPPIAIAWLPGED---------TDDGTLKFGGWSWRYDLESL----APA
TH_4053|M.thermoresistible__bu FDPPHAIAWQPGAEPRHIPGRGADATGPVEFGGWIWRYDLAPQHAPGAPA
MMAR_0253|M.marinum_M FTEARLIAWRPSELG-------------KTPPGHLWRWELTPIS----GT
MUL_4861|M.ulcerans_Agy99 FTEARLIAWRPSELG-------------KTPPGHLWRWELTPIS----GT
MAB_2457c|M.abscessus_ATCC_199 FEEARRIAWLPSEVG-------------KRPPGHLWRWELRPLD----DD
MAV_5230|M.avium_104 FVEGSVIAWRPAEPG-------------KQPPGHLWRWELSPVN----AE
Mb0091|M.bovis_AF2122/97 LKPNELVEWS-------------------HPLGHRWRWEFESLS----PT
Rv0088|M.tuberculosis_H37Rv LKPNELVEWS-------------------HPLGHRWRWEFESLS----PT
: : * * *:: : .
Mflv_0146|M.gilvum_PYR-GCK QTRVTLTYDWSGVPAELR-----EHIEFPPFPDTHLKNSLDNLATLVVG-
Mvan_0758|M.vanbaalenii_PYR-1 LTRVTLTYDWSAVPATLR-----EHIAFPPFPVDHLHNSLANLAKLAAG-
MSMEG_3417|M.smegmatis_MC2_155 RTRITLTYDWSAVPDAIR-----QHIGFPPFDPQHLDNSLKHLAGLAEKG
TH_4053|M.thermoresistible__bu GTRVTLTYDWSQVPAADR-----RAMRFPPFPPEHLVNSLRHLKELAER-
MMAR_0253|M.marinum_M RTLVTHTYDWTELSDPA------RFPRARATTTVQLRASLDRLAGHAEAA
MUL_4861|M.ulcerans_Agy99 RTLVTHTYDWTELSDPA------RFPRARATTTVQLRASLDRLAGHAEAA
MAB_2457c|M.abscessus_ATCC_199 RTQVTHTYDWTNLTDEK------RLVRARNTTSERLQASVDRLAALAESG
MAV_5230|M.avium_104 LTTVTHTYDWTALTDPT------RLQRARATTPEMLRESMDRLATLAQSG
Mb0091|M.bovis_AF2122/97 LTRVTETFDYHAAGAIKNGLKFYEMTGFAKSNAAGIEATLAKLSDQYARG
Rv0088|M.tuberculosis_H37Rv LTRVTETFDYHAAGAIKNGLKFYEMTGFAKSNAAGIEATLAKLSDQYARG
* :* *:*: . : :: .*
Mflv_0146|M.gilvum_PYR-GCK ---------
Mvan_0758|M.vanbaalenii_PYR-1 ---------
MSMEG_3417|M.smegmatis_MC2_155 SAEVDVSSG
TH_4053|M.thermoresistible__bu ----DVPR-
MMAR_0253|M.marinum_M AAGD-----
MUL_4861|M.ulcerans_Agy99 AAGD-----
MAB_2457c|M.abscessus_ATCC_199 A--------
MAV_5230|M.avium_104 D--------
Mb0091|M.bovis_AF2122/97 RA-------
Rv0088|M.tuberculosis_H37Rv RA-------