For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLNQIVVAGAIVRGCTVLVAQRVRPPELAGRWELPGGKVAAGETERAALARELAEELGLEVADLAVGDRV GDDIALNGTTTLRAYRVHLLGGEPRARDHRALCWVTAAELHDVDWVPADRGWIADLARTLNGSAADVHRR C
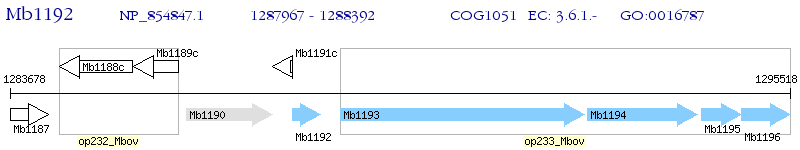
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1192 | mutT2 | - | 100% (141) | 7,8-dihydro-8-oxoguanine-triphosphatase |
| M. bovis AF2122 / 97 | Mb3938 | - | 1e-05 | 32.58% (89) | hypothetical protein Mb3938 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2154 | - | 6e-37 | 57.89% (133) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv1160 | mutT2 | 4e-79 | 100.00% (141) | 7,8-dihydro-8-oxoguanine-triphosphatase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1308 | - | 2e-31 | 54.20% (131) | mutator protein MutT2/NUDIX hydrolase |
| M. marinum M | MMAR_4290 | mutT2 | 4e-51 | 76.56% (128) | mutator protein MutT2 |
| M. avium 104 | MAV_1301 | - | 1e-54 | 76.92% (130) | hydrolase, NUDIX family protein |
| M. smegmatis MC2 155 | MSMEG_5148 | - | 2e-39 | 60.61% (132) | CTP pyrophosphohydrolase |
| M. thermoresistible (build 8) | TH_0645 | mutT2 | 2e-42 | 64.84% (128) | PROBABLE MUTATOR PROTEIN MUTT |
| M. ulcerans Agy99 | MUL_1008 | mutT2 | 4e-49 | 74.22% (128) | mutator protein MutT2 |
| M. vanbaalenii PYR-1 | Mvan_4548 | - | 5e-36 | 58.14% (129) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1192|M.bovis_AF2122/97 --------------------------------------------------
Rv1160|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1301|M.avium_104 --------------------------------------------------
MMAR_4290|M.marinum_M -------MRAIMFGLSKPAWRAVKAAVAQGWGVLGQHGFGRIAVRRIWEV
MUL_1008|M.ulcerans_Agy99 --------------------------------------------------
MAB_1308|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2154|M.gilvum_PYR-GCK MVFALGVLADAPDRVDAVEKRGELDRSSQRPVGALPTVELGQNGVHLFVS
Mvan_4548|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5148|M.smegmatis_MC2_155 --------------------------------------------------
TH_0645|M.thermoresistible__bu --------------------------------------------------
Mb1192|M.bovis_AF2122/97 -------------MLN---QIVVAGAIVRGCTVLVAQRVRPPELAGRWEL
Rv1160|M.tuberculosis_H37Rv -------------MLN---QIVVAGAIVRGCTVLVAQRVRPPELAGRWEL
MAV_1301|M.avium_104 ---MRADGIPSLAMPT---QIVVAGALIRGSRLLVAQRARPPELAGRWEL
MMAR_4290|M.marinum_M ALPGRLGEIPSLAMPNQPNQIVVAGAIIRGATVLVAQRARPPELAGRWEL
MUL_1008|M.ulcerans_Agy99 -------------MPNQPNQIVVAGAFICGATVLVAQRVRPPELAGRWEL
MAB_1308|M.abscessus_ATCC_1997 ---------------------MVAGAVIDGDKLLIAQRAKPAELAGQWEL
Mflv_2154|M.gilvum_PYR-GCK QYSHAFDGIPSSPMSN---LIVVAGALIEHGALLVAQRARPPELAGLWEL
Mvan_4548|M.vanbaalenii_PYR-1 -------------MSK---LIVVAGALIEGGALLVAQRDRPPELAGLWEL
MSMEG_5148|M.smegmatis_MC2_155 -------------MTK---QIVVAGALISRGTLLVAQRDRPAELAGLWEL
TH_0645|M.thermoresistible__bu ---------MTQIPQP---QIVVAGALIADAALLVAQRNRPPELAGRWEL
:****.: :*:*** :*.**** ***
Mb1192|M.bovis_AF2122/97 PGGKVAAGETERAALARELAEELGLEVADLAVGDRVGDDIALNGTTTLRA
Rv1160|M.tuberculosis_H37Rv PGGKVAAGETERAALARELAEELGLEVADLAVGDRVGDDIALNGTTTLRA
MAV_1301|M.avium_104 PGGKVAPGETERDALARELAEELGLRVGDIAVGDRLGGDIAVDGGMTLRA
MMAR_4290|M.marinum_M PGGKVAGGETEPAALARELVEELGLGVDDVAVGERLGADVVLDEKTVLRA
MUL_1008|M.ulcerans_Agy99 PGGKVAGGETEPAALARELVEELGLGVDDVAVGERLGADVVPDEKTVLRA
MAB_1308|M.abscessus_ATCC_1997 PGGKVADGESEPQALVRELREELGIEV---EVGARLGEDVVVG-NLVLRA
Mflv_2154|M.gilvum_PYR-GCK PGGKVTPPESDESALARELNEELGIDV---TVGPRIGADVALSATTTLRA
Mvan_4548|M.vanbaalenii_PYR-1 PGGKVAPGESDEAALVRELNEELGVDV---TVGARVGADIALSAAMCLRA
MSMEG_5148|M.smegmatis_MC2_155 PGGKVTPGESDADALARELREELGVDV---AVGERLGADVALNDAMTLRA
TH_0645|M.thermoresistible__bu PGGKVAPGETEAEALVRELKEELGVDV---EVGARLGVEVALTESMVLRA
*****: *:: **.*** ****: * ** *:* ::. ***
Mb1192|M.bovis_AF2122/97 YRVHLLG---GEPRARDHRALCWVTAAELHDVDWVPADRGWIADLARTLN
Rv1160|M.tuberculosis_H37Rv YRVHLLG---GEPRARDHRALCWVTAAELHDVDWVPADRGWIADLARTLN
MAV_1301|M.avium_104 YRVRLLR---GRPDARDHRALRWITAAQLHDLDWVPADRGWLGDLARVL-
MMAR_4290|M.marinum_M YRVRLLR---GHPCARDHRALRWVTADQLDDVDWVPADRCWLADLDRALR
MUL_1008|M.ulcerans_Agy99 YRVRLLR---GHPCARDHRALRWVTVDQLDDVDWVPADRCWLADLDRALR
MAB_1308|M.abscessus_ATCC_1997 YSARLHPQHPGSPHPHEHLALRWVTAGELDAVEWVAADGGWIPALKQALV
Mflv_2154|M.gilvum_PYR-GCK YVVTRTR---GAVTAHDHRALRWVRADEIPSLPWVPADRAWLPDLVGLLA
Mvan_4548|M.vanbaalenii_PYR-1 YAVTRTR---GVVAPHDHRALRWIRTEEIETLPWVPADRAWLPDLTRLLA
MSMEG_5148|M.smegmatis_MC2_155 YRVTLRS---GSPHPHDHRALRWVGADEIDGLAWVPADRAWVPDLVAALS
TH_0645|M.thermoresistible__bu FRVHHRG---GSPSPNDHRALRWVTAAELHDLDWVPADRAWIPELTALLS
: . * ..:* ** *: . :: : **.** *: * *
Mb1192|M.bovis_AF2122/97 GSAADVHRRC----------
Rv1160|M.tuberculosis_H37Rv GSAADVHRRC----------
MAV_1301|M.avium_104 --------------------
MMAR_4290|M.marinum_M PAPPPG--------------
MUL_1008|M.ulcerans_Agy99 PAPPTG--------------
MAB_1308|M.abscessus_ATCC_1997 GTRPV---------------
Mflv_2154|M.gilvum_PYR-GCK GNNLTQSGSGTDTDG-----
Mvan_4548|M.vanbaalenii_PYR-1 GADHARASGAAARRGNQPPA
MSMEG_5148|M.smegmatis_MC2_155 GR------------------
TH_0645|M.thermoresistible__bu AR------------------