For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSLAVTMFKRARAEIFDRNREVGISNVTTAASLVTFPVLAGILGGVVPSVRTPSAAMVSGVQHFAAGIVM AAVAGEVLPDLRSRGPLWLIVVGFSAGVAVLVALRRFDGHGEHQDGDDVGELPVGFLTVVAVDLFIDGLL VATGATVSSRTAIIITIALTVEVLFLGLAVALRLAGSGMPRIRAAATTSALSLVIAVGGVSGAVALGRAG NTVLTLVLAFAAAALLWLVVEELLVEAHETPERPWMAVMFFAGFLILYGLGVME
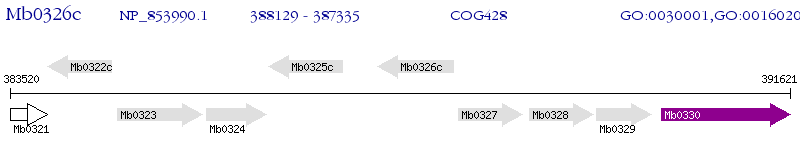
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0326c | - | - | 100% (264) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv0318c | - | 1e-142 | 99.62% (264) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0595 | - | 4e-98 | 75.50% (249) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5407 | - | 3e-06 | 28.26% (230) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0554 | - | 2e-99 | 76.31% (249) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0326c|M.bovis_AF2122/97 MSLAVTMFKRARAEIFDRNREVGISNVTTAASLVTFPVLAGILGGVVPSV
Rv0318c|M.tuberculosis_H37Rv MSLAVTMFKRARAEIFDRNREVGISNVTTAASLVTFPVLAGILGGVVPSV
MMAR_0595|M.marinum_M ------MFVSIGPGRF------GVSTLTTAAALVAFPVIAGIIGGAVAVV
MUL_0554|M.ulcerans_Agy99 ------MFVSIGPGRF------GVSTLTTAAALVAFPVIAGIIGGAVAVV
MSMEG_5407|M.smegmatis_MC2_155 --------------------------MLTALLWGLVAASSLLVGAVAGTV
: ** ... : ::*... *
Mb0326c|M.bovis_AF2122/97 RTPSAAMVSGVQHFAAGIVMAAVAGEVLPDLRSRGPLWLIVVGFSAG-VA
Rv0318c|M.tuberculosis_H37Rv RTPSAAMVSGVQHFAAGIVMAAVAGEVLPDLRSRGPLWLIVVGFSAG-VA
MMAR_0595|M.marinum_M RTPSTALVSGVQHFAAGVVMAAVATEVLPDLRARGPLWLIILGFSAG-VA
MUL_0554|M.ulcerans_Agy99 RTPSAALVSGVQHFAAGVVMAAVATEVLPDLRARGPLWLIILGFSAG-VA
MSMEG_5407|M.smegmatis_MC2_155 RDWNERLVGLVLGFGAGALISSISFELAEEGVRVSGALAVALGLAIGAVV
* . :*. * *.** :::::: *: : . : :*:: * *.
Mb0326c|M.bovis_AF2122/97 VLVALRRFDGHG---EHQDG-DDV---GELPVGFLTVVAVDLFIDGLLVA
Rv0318c|M.tuberculosis_H37Rv VLVALRRFDGHG---EHQDG-DDV---GELPVGFLTVVAVDLFIDGLLVA
MMAR_0595|M.marinum_M LLAGFRQFEDHGPHGEDEDGHDDVRAGGQVPVGFLAVVAVDLFIDGLLVA
MUL_0554|M.ulcerans_Agy99 LLVGFRQFEDHGPHGEDEDGHDDVRAGGQVPVGFLAVVAVDLFIDGLLVA
MSMEG_5407|M.smegmatis_MC2_155 FYVADKTVDRMG-----RTG-------GGAGLPLLLGALLDGIPEQAVLG
. .. : .: * . * * : :* . :* : : ::.
Mb0326c|M.bovis_AF2122/97 TGATVSSRTAIIITIALTVEVLFLGLAVALRLAGSGMPRIRAAATTSALS
Rv0318c|M.tuberculosis_H37Rv TGATVSSRTAIIITIALTVEVLFLGLAVALRLAGSGMPRIRAAATTSALS
MMAR_0595|M.marinum_M TGATVSRRTAIIITIALTVEVLFLGVSVALRLTRSGVPKARAAALTGGVS
MUL_0554|M.ulcerans_Agy99 TGATVSRRTAIIITIALTVEVLFLGVSVALRLTRSGVPKARAAALTGGVS
MSMEG_5407|M.smegmatis_MC2_155 IGIAGGAGVSMALLVAIFVSNLPESIGSASDMRSAGHPVRTILGGWAAIS
* : . .:: : :*: *. * .:. * : :* * . ..:*
Mb0326c|M.bovis_AF2122/97 LVIAVGGVSGAVALGRAGNTVLTLVLAFAAAALLWLVVEELLVEAHETPE
Rv0318c|M.tuberculosis_H37Rv LVIAVGGVSGAVALGRAGNTVLTLVLAFAAGALLWLVVEELLVEAHETPE
MMAR_0595|M.marinum_M LATAVGGVLGALVLANAGGTVLTLVLAFAAAALLWLVVEELLVEAHEGEE
MUL_0554|M.ulcerans_Agy99 LATAVGGVPGALVLANAGGTVLTLVLAFAAAALLWLVVEELLVEAHEGEE
MSMEG_5407|M.smegmatis_MC2_155 ALCAVATVGGYQLQKVAGAALQGGINGFAAGALLVMLVGSMIPEATQK-A
**. * * ** :: : .***.*** ::* .:: ** :
Mb0326c|M.bovis_AF2122/97 RPWMAVMFFAGFLILYGLGVME
Rv0318c|M.tuberculosis_H37Rv RPWMAVMFFAGFLILYGLGVME
MMAR_0595|M.marinum_M RAWMAIMFFAGFLILYSLGVME
MUL_0554|M.ulcerans_Agy99 RAWMAIMFFAGFLILYSLGVME
MSMEG_5407|M.smegmatis_MC2_155 RENAGLAAVLGFAVAAGLSLAT
* .: . ** : .*.: