For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFSAPVPTTPTRLTGWGRTAPSVAEVLRTPDPEVIATAVARVADTGARGVIARGLGRSYGDNAQNGGGLV IDMSPLNKIHSISADSTLVDVDAGVNLDQLMKAALPLGLWVPVLPGTRQVTIGGAIACDIHGKNHHSAGS FGNHVRSLDLLTADGEVRHLTPTGTETADTELFWATVGGNGLTGIILRATIEMTPTETAYFIADGDVTAS LDETIALHSDGSGGNYTYSSAWFDAISGPPKLGRAAVSRGCLATIDQLPTKLQRDPLKFDAPQLLTFPDV FPNGLANKYTFGPIGELWYRKSGTYRGKVQNLTQFYHPLDMFGEWNRAYGPAGFLQYQFVVPTEAVDEFK RIIGDIQASGHYSFLNVFKLFGAGNQAPLSFPIPGWNICVDFPIKAGLNEFVSELDRRVMEFGGRLYTAK DSRTTAETFHAMYPRIDEWIAVRRKVDPCGVFASDMARRLELL
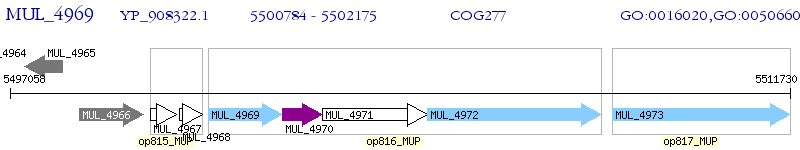
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4969 | - | - | 100% (463) | oxidoreductase |
| M. ulcerans Agy99 | MUL_4485 | - | 2e-07 | 27.00% (200) | oxidoreductase |
| M. ulcerans Agy99 | MUL_1431 | - | 5e-07 | 30.19% (159) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3819 | - | 0.0 | 87.50% (464) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1178 | - | 0.0 | 85.31% (463) | FAD linked oxidase domain-containing protein |
| M. tuberculosis H37Rv | Rv3790 | - | 0.0 | 87.50% (464) | oxidoreductase |
| M. leprae Br4923 | MLBr_00109 | - | 0.0 | 84.88% (463) | putative FAD-linked oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0192c | - | 0.0 | 68.82% (465) | oxidoreductase |
| M. marinum M | MMAR_5352 | - | 0.0 | 99.14% (463) | oxidoreductase |
| M. avium 104 | MAV_0232 | - | 0.0 | 77.29% (458) | oxidoreductase, FAD-binding |
| M. smegmatis MC2 155 | MSMEG_6382 | - | 0.0 | 84.45% (463) | oxidoreductase, FAD-binding |
| M. thermoresistible (build 8) | TH_1603 | - | 0.0 | 84.12% (466) | PROBABLE OXIDOREDUCTASE |
| M. vanbaalenii PYR-1 | Mvan_5627 | - | 0.0 | 81.76% (466) | FAD linked oxidase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4969|M.ulcerans_Agy99 --------------------------------------MFS--APVPTTP
MMAR_5352|M.marinum_M --------------------------------------MFS--APVPTTP
Mb3819|M.bovis_AF2122/97 --------------------------------------MLS--VGATTTA
Rv3790|M.tuberculosis_H37Rv --------------------------------------MLS--VGATTTA
MLBr_00109|M.leprae_Br4923 --------------------------------------MLN--MGVSTTA
Mflv_1178|M.gilvum_PYR-GCK --------------------------MSTGPQAVPSVTMLS--TEFPTTA
Mvan_5627|M.vanbaalenii_PYR-1 --------------------------------------MLS--TEFPTTA
MSMEG_6382|M.smegmatis_MC2_155 ------------------------------MGAVPSLTMST--TEFPTTT
TH_1603|M.thermoresistible__bu --------------------------------------MRT--TEVETTP
MAB_0192c|M.abscessus_ATCC_199 MARASGLCHRPGDCDSHQFHRPARGDLPNPLSRYAQLPMTTPKSELPLTP
MAV_0232|M.avium_104 --------------------------------------MSS--TDPLITP
* . *.
MUL_4969|M.ulcerans_Agy99 TRLTGWGRTAPSVAEVLRTPDPEVIATAVARVADTGA-------RGVIAR
MMAR_5352|M.marinum_M TRLTGWGRTAPSVAEVLRTPDPEVIATAVARVADSGA-------RGVIAR
Mb3819|M.bovis_AF2122/97 TRLTGWGRTAPSVANVLRTPDAEMIVKAVARVAESGGG------RGAIAR
Rv3790|M.tuberculosis_H37Rv TRLTGWGRTAPSVANVLRTPDAEMIVKAVARVAESGGG------RGAIAR
MLBr_00109|M.leprae_Br4923 TRLTGWGRTAPSVANVLSTPDAELISKVVARVADADG-------RGVIAR
Mflv_1178|M.gilvum_PYR-GCK QRLTGWGRTAPSVAQVLSTPDPEVIVKAVTRAAENP-------GRGVLAR
Mvan_5627|M.vanbaalenii_PYR-1 RRLTGWGRTAPSVARVLATPDPEVIVKAVARAAESGKG----GGRGVIAR
MSMEG_6382|M.smegmatis_MC2_155 KRLMGWGRTAPTVASVLSTSDPEVIVRAVTRAAEEG-------GRGVIAR
TH_1603|M.thermoresistible__bu RRLTGWGRTAPTVAQVLSTSDAEVIAEAVRRTAEQSKA----GARGVIAR
MAB_0192c|M.abscessus_ATCC_199 RALTGFGRTAPTVAHVLSTPDVDVIAEAVRQVADASSHSPAHLRRGIVAR
MAV_0232|M.avium_104 ARLTGFGRTAPSVAQVLRTRDPEVIAKAVARVADSG-HSKS---RGVIAR
* *:*****:** ** * * ::* .* :.*: ** :**
MUL_4969|M.ulcerans_Agy99 GLGRSYGDNAQNGGGLVIDMSPLNKIHSISADSTLVDVDAGVNLDQLMKA
MMAR_5352|M.marinum_M GLGRSYGDNAQNGGGLVIDMSPLNKIHSISADSKLVDVDAGVNLDQLMKA
Mb3819|M.bovis_AF2122/97 GLGRSYGDNAQNGGGLVIDMTPLNTIHSIDADTKLVDIDAGVNLDQLMKA
Rv3790|M.tuberculosis_H37Rv GLGRSYGDNAQNGGGLVIDMTPLNTIHSIDADTKLVDIDAGVNLDQLMKA
MLBr_00109|M.leprae_Br4923 GLGRSYGDNAQNGGGLVIDMTPLNTIHCINTDTKLADVDAGVNLNKLMKA
Mflv_1178|M.gilvum_PYR-GCK GLGRSYGDNAQNGGGLVIDMSSLDRIHSMDADSRLVDVDGGVNLDQLMRA
Mvan_5627|M.vanbaalenii_PYR-1 GLGRSYGDNAQNGGGLVVDMSVLNRIHSMDSDSHLVDVDGGVNLDQLMRA
MSMEG_6382|M.smegmatis_MC2_155 GLGRSYGDNAQNGGGLVIDMPALNRIHSIDSGTRLVDVDAGVSLDQLMKA
TH_1603|M.thermoresistible__bu GLGRSYGDNAQNGGGLVIDMTPLNRIHRIDAETRLVDLDAGVSLDTLMKA
MAB_0192c|M.abscessus_ATCC_199 GLGRSYGDHACNGGGIVVDMTPLKRVHSISAETAVADVDAGVSLDQLMKA
MAV_0232|M.avium_104 GLGRSYGDNAQNGGGLVIDMTGLNRIHSISADTRLVDVDAGVSLDQLMKA
********:* ****:*:**. *. :* :.: : :.*:*.**.*: **:*
MUL_4969|M.ulcerans_Agy99 ALPLGLWVPVLPGTRQVTIGGAIACDIHGKNHHSAGSFGNHVRSLDLLTA
MMAR_5352|M.marinum_M ALPLGLWVPVLPGTRQVTIGGAIACDIHGKNHHSAGSFGNHVRSLDLLTA
Mb3819|M.bovis_AF2122/97 ALPFGLWVPVLPGTRQVTVGGAIACDIHGKNHHSAGSFGNHVRSMDLLTA
Rv3790|M.tuberculosis_H37Rv ALPFGLWVPVLPGTRQVTVGGAIACDIHGKNHHSAGSFGNHVRSMDLLTA
MLBr_00109|M.leprae_Br4923 ALPFGLWIPVLPGTRQVTIGGAIACDIHGKNHHSAGSFGNYVRSMDLLTA
Mflv_1178|M.gilvum_PYR-GCK ALPLGLWVPVLPGTRQVTIGGAIACDIHGKNHHSAGSFGNHVRSIDLLTA
Mvan_5627|M.vanbaalenii_PYR-1 ALPLGLWVPVLPGTRQVTVGGAIACDIHGKNHHSAGSFGNHVRSMDLLTA
MSMEG_6382|M.smegmatis_MC2_155 ALPHGLWVPVLPGTRQVTVGGAIGCDIHGKNHHSAGSFGNHVRSMELLTA
TH_1603|M.thermoresistible__bu ALPFGLWVPVLPGTRQVTVGGAIACDIHGKNHHSAGSFGNHVRSIDLLTA
MAB_0192c|M.abscessus_ATCC_199 ALPFGLWVPVLPGTRQVTVGGAIGSDIHGKNHHSAGSFGNHVLSMDLLMA
MAV_0232|M.avium_104 ALPFGLWVPVLPGTRQVTVGGAIACDIHGKNHHSAGSFGNHVRSMELLMA
*** ***:**********:****..***************:* *::** *
MUL_4969|M.ulcerans_Agy99 DGEVRHLTPTGTETADTELFWATVGGNGLTGIILRATIEMTPTETAYFIA
MMAR_5352|M.marinum_M DGEVRHLTPTGTETADTALFWATVGGNGLTGIILRATIEMTPTETAYFIA
Mb3819|M.bovis_AF2122/97 DGEIRHLTPTG---EDAELFWATVGGNGLTGIIMRATIEMTPTSTAYFIA
Rv3790|M.tuberculosis_H37Rv DGEIRHLTPTG---EDAELFWATVGGNGLTGIIMRATIEMTPTSTAYFIA
MLBr_00109|M.leprae_Br4923 SGEVRRLTPTG---KDSELFWATVGGNGLTGIIMRATIEMMPTETAYFIA
Mflv_1178|M.gilvum_PYR-GCK DGRVRTITPDG---PESPLFWATVGGNGLTGIILRATIEMTPTETAYFIA
Mvan_5627|M.vanbaalenii_PYR-1 DGEVRTLTPSG---PDADLFWATVGGNGLTGIILRATIEMTPTETAYFIA
MSMEG_6382|M.smegmatis_MC2_155 NGEVRHLTPAG---PDSDLFWATVGGNGLTGIILRATIEMTPTETAYFIA
TH_1603|M.thermoresistible__bu DGQIRRLTPDG---DDPELFWATVGGNGLTGIILRATIEMTPTETAYFIA
MAB_0192c|M.abscessus_ATCC_199 DGEVHTITPDGP---ASELFWATVGGNGLTGIVVRARIAMTRTETAYFIA
MAV_0232|M.avium_104 DGTVRTITPDGPDASDAELFWATVGGNGLTGIVLRATIAMTPTETAYFIA
.* :: :** * . **************::** * * *.******
MUL_4969|M.ulcerans_Agy99 DGDVTASLDETIALHSDGSGGNYTYSSAWFDAISGPPKLGRAAVSRGCLA
MMAR_5352|M.marinum_M DGDVTASLDETIALHSDGSEGNYTYSSAWFDAISGPPKLGRAAVSRGCLA
Mb3819|M.bovis_AF2122/97 DGDVTASLDETIALHSDGSEARYTYSSAWFDAISAPPKLGRAAVSRGRLA
Rv3790|M.tuberculosis_H37Rv DGDVTASLDETIALHSDGSEARYTYSSAWFDAISAPPKLGRAAVSRGRLA
MLBr_00109|M.leprae_Br4923 DGDITGSLDETIALHSDGSEADYTYSSAWFDAISASPKLGRAAVSRGCLA
Mflv_1178|M.gilvum_PYR-GCK DGDVTSGLDETIAFHSDGTEDNYTYSSAWFDAISPPPKLGRAAISRGSLA
Mvan_5627|M.vanbaalenii_PYR-1 DGDVTSGLEETIAFHSDGTEDNYTYSSAWFDAISAPPKLGRAVISRGSLA
MSMEG_6382|M.smegmatis_MC2_155 DGDVTGSLDETIAFHSDGSEANYTYSSAWFDAISKPPKLGRAAISRGSLA
TH_1603|M.thermoresistible__bu DGDVTATLDETIALHSDGSEDNYTYSSAWFDAISAPPKLGRAAISRGSLA
MAB_0192c|M.abscessus_ATCC_199 DGVATRDLDETIAVHQDGTEDNYTYSSAWFDLINPPPKLGRAAVSRGSLA
MAV_0232|M.avium_104 DGMATKDLDETVAVHLDGSEADYTYSSAWFDLISPPPKLGRAAVSRGSLA
** * *:**:*.* **: ********* *. .******.:*** **
MUL_4969|M.ulcerans_Agy99 TIDQLPTKLQRDPLKFDAPQLLTFPDVFPNGLANKYTFGPIGELWYRKSG
MMAR_5352|M.marinum_M TIDQLPTKLQRDPLKFDAPQLLTFPDVFPNGLANKYTFGPIGELWYRKSG
Mb3819|M.bovis_AF2122/97 TVEQLPAKLRSEPLKFDAPQLLTLPDVFPNGLANKYTFGPIGELWYRKSG
Rv3790|M.tuberculosis_H37Rv TVEQLPAKLRSEPLKFDAPQLLTLPDVFPNGLANKYTFGPIGELWYRKSG
MLBr_00109|M.leprae_Br4923 RLDQLPAKLRRNPLKFHAPQLLTLPDVFPNGLANKYTFMPIGELWYRKSG
Mflv_1178|M.gilvum_PYR-GCK TVDQLPKKLQKNPLKFDAPQLLTFPDLFPNGLANKYTFGPIGELWYRKSG
Mvan_5627|M.vanbaalenii_PYR-1 TVDQLPRKLQKNPLKFDAPQYFTAPEIFPNGLGNKYTFGPMTEVWYRLGK
MSMEG_6382|M.smegmatis_MC2_155 KLDQLPSKLQKDPLKFDAPQLLTLPDIFPNGLANKFTFMPIGELWYRKSG
TH_1603|M.thermoresistible__bu RLDELPPKLRKNPLKFDAPQLLTFPDIFPNGLANKYTFGPIGELWYRKSG
MAB_0192c|M.abscessus_ATCC_199 KLDQLPPKLAKDPLKFSAPQLPGLPDLFPVNFMIKPSLMAIGEAFYRMSG
MAV_0232|M.avium_104 RLDQLPKKLAKNPLKFDAPQLLTVPDVFPVSAMNKLSFMAIGEVYYRLGG
:::** ** :**** *** *::** . * :: .: * :** .
MUL_4969|M.ulcerans_Agy99 TYRGKVQNLTQFYHPLDMFGEWNRAYGPAGFLQYQFVVPTEAVDEFKRII
MMAR_5352|M.marinum_M TYRGKVQNLTQFYHPLDMFGEWNRAYGPAGFLQYQFVVPTEAVDEFKRII
Mb3819|M.bovis_AF2122/97 TYRGKVQNLTQFYHPLDMFGEWNRAYGPAGFLQYQFVIPTEAVDEFKKII
Rv3790|M.tuberculosis_H37Rv TYRGKVQNLTQFYHPLDMFGEWNRAYGPAGFLQYQFVIPTEAVDEFKKII
MLBr_00109|M.leprae_Br4923 TYRGKIQNLTQFYHPLDMFGEWNRAYGAAGFLQYQFVIPTEAVDKVKRII
Mflv_1178|M.gilvum_PYR-GCK TYRGKVQNLTQFYHPLDMFGEWNRAYGAAGFTQYQFVVPTTAVDEFKAII
Mvan_5627|M.vanbaalenii_PYR-1 TYRGKAQNLTQFYHPLDMVGDWNRAYGSVGFAQYQFVVPTEAVDEFKRIM
MSMEG_6382|M.smegmatis_MC2_155 TYRNKVQNLTQFYHPLDMFGEWNRAYGSAGFLQYQFVVPTEAVEEFKSII
TH_1603|M.thermoresistible__bu TYRNKIQNLTQFYHPLDMFGEWNRAYGPAGFTQYQFVVPTEAVDEFKAIL
MAB_0192c|M.abscessus_ATCC_199 NYQGKIVNLTQFYHMLDITTGWQYAYGPAGFAQHQFLVPPDALEEFKGII
MAV_0232|M.avium_104 TYTGKIMNLSQFYHMLDLVSGWNNAYGPRGFAQHQFLVPPDAMDEFKAII
.* .* **:**** **: *: ***. ** *:**::*. *:::.* *:
MUL_4969|M.ulcerans_Agy99 GDIQASGHYSFLNVFKLFGAGNQAPLSFPIPGWNICVDFPIKAGLNEFVS
MMAR_5352|M.marinum_M GDIQASGHYSFLNVFKLFGAGNQAPLSFPIPGWNICVDFPIKAGLNEFVS
Mb3819|M.bovis_AF2122/97 GVIQASGHYSFLNVFKLFGPRNQAPLSFPIPGWNICVDFPIKDGLGKFVS
Rv3790|M.tuberculosis_H37Rv GVIQASGHYSFLNVFKLFGPRNQAPLSFPIPGWNICVDFPIKDGLGKFVS
MLBr_00109|M.leprae_Br4923 GDMQGSGHYSFLNVFKLFGPGNQAPLSFPIPGWNICVDFPIKSGLNEFVN
Mflv_1178|M.gilvum_PYR-GCK VDIQRSGFYSFLNVFKLFGPGNQAPLSFPIPGWNVCVDFPITAGLNEFLN
Mvan_5627|M.vanbaalenii_PYR-1 YDIQRSGHYSFLNVFKLFGPGNDAPLSFPIPGWNVCVDFQINPGLNEFLN
MSMEG_6382|M.smegmatis_MC2_155 VDIQRSGHYSFLNVFKLFGPGNQAPLSFPIPGWNVCVDFPIKAGLHEFVT
TH_1603|M.thermoresistible__bu VDIQRSGHYSFLNVFKLFGPGNKAPLSFPIPGWNVCVDFPVKPGLNEFLT
MAB_0192c|M.abscessus_ATCC_199 RWIQTQGQYSALNVFKLFGPGNKAPLSFPMKGWNVAMDFPNKPGVNEFLN
MAV_0232|M.avium_104 RWIQTRGHYSALNVFKLFGPGNRAPLSFPMAGWNVAMDFPNKPGVNEFLN
:* * ** ********. * ******: ***:.:** . *: :*:.
MUL_4969|M.ulcerans_Agy99 ELDRRVMEFGGRLYTAKDSRTTAETFHAMYPRIDEWIAVRRKVDPCGVFA
MMAR_5352|M.marinum_M ELDRRVMEFGGRLYTAKDSRTTAETFHAMYPRIDEWIAVRRKVDPCGVFA
Mb3819|M.bovis_AF2122/97 ELDRRVLEFGGRLYTAKDSRTTAETFHAMYPRVDEWISVRRKVDPLRVFA
Rv3790|M.tuberculosis_H37Rv ELDRRVLEFGGRLYTAKDSRTTAETFHAMYPRVDEWISVRRKVDPLRVFA
MLBr_00109|M.leprae_Br4923 KLDRRVMELGGRLYTAKDSRTTADTFHAMYPRIDEWIAVRRKVDPLRVFS
Mflv_1178|M.gilvum_PYR-GCK GLDKRVLQFGGRLYTAKDSRTTAETFHAMYPRIDEWIAVRRNVDPDGVFA
Mvan_5627|M.vanbaalenii_PYR-1 GLDKRVLEFGGRLYTAKDSRTTAETFHAMYPRIDEWIALRRKVDPDGVFA
MSMEG_6382|M.smegmatis_MC2_155 ELDRRVLEFGGRLYTAKDSRTTAETFHAMYPRIDEWIRIRRSVDPDGVFA
TH_1603|M.thermoresistible__bu GLDRRVLEFGGRLYTAKDSRTTAETFHAMYPRIDEWIAVRRRVDPEGIFA
MAB_0192c|M.abscessus_ATCC_199 ELDNRAMEFGGRVYTAKDSRVSAEKFHRMYPRVDEWIATRRKADPHGVFA
MAV_0232|M.avium_104 ELDRRVLQFGGRVYTAKDSRTNAETFHAMYPRIDEWIAVRRKVDPTGVFA
**.*.:::***:*******..*:.** ****:**** ** .** :*:
MUL_4969|M.ulcerans_Agy99 SDMARRLELL
MMAR_5352|M.marinum_M SDMARRLELL
Mb3819|M.bovis_AF2122/97 SDMARRLELL
Rv3790|M.tuberculosis_H37Rv SDMARRLELL
MLBr_00109|M.leprae_Br4923 SDMARRLELL
Mflv_1178|M.gilvum_PYR-GCK SDMARRLELL
Mvan_5627|M.vanbaalenii_PYR-1 SDMARRLELL
MSMEG_6382|M.smegmatis_MC2_155 SDMARRLQLL
TH_1603|M.thermoresistible__bu SDMARRLELL
MAB_0192c|M.abscessus_ATCC_199 SDMARRLELL
MAV_0232|M.avium_104 SDMARRLELL
*******:**