For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRDVSDFAGEFVSGSGTAAYCDAITLASGLVANALEAADAPGLVDGYSLLKELADKPRLDTVAGVGLPAA LEEAVGLMAGHSAVVTHPAYLAHLHCPPTVASLSAEVLISAFNQSLDSFDQAPAATAIEQRVVEHLCARI GYGTDADGTFTSGGTQSNLHALLMARDVFAQRTLGLDVGTAGLPAAARSWRLLCTRQAHFSARQALRILG LGADAVIEVPTDDAGRLCVEALAGCIAESQARGTPVFGLFLTAGTTDYGAIDPLEPAIKIARQHGVWAHV DACVGGCLAFSAQHRHLLRGIELADSVAVDFHKLLFQAISCSALLVRDAESFRVSAGHADYLNPADDVEH DVVNLVGKSLQTTRRFDALKIFLTLRALGERRIGAMIDATCAAAAAAGRAVSDHPHLTLCAPVAVNTVVL RWRHPELTEQKCDAVNSAIRSSLARCGLALVGRSRAAGHQAIKLTFVNPLITADLATGVIADVSRRGNEI AGLPDDLAAQA*
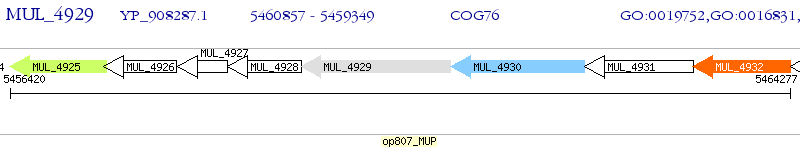
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4929 | - | - | 100% (502) | glutamate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1685 | - | 0.0 | 66.05% (486) | putative decarboxylase |
| M. marinum M | MMAR_0167 | - | 0.0 | 99.60% (502) | glutamate decarboxylase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3506 | - | 3e-37 | 32.68% (355) | putative amino acid decarboxylase, pyridoxal-dependent protein |
| M. thermoresistible (build 8) | TH_2178 | - | 3e-40 | 30.60% (464) | putative amino acid decarboxylase, Pyridoxal-dependent protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4929|M.ulcerans_Agy99 MSRDVS-----DFAGEFVSGSGTAAYCDAITLASGLVANALEAADAPGLV
MMAR_0167|M.marinum_M MSRDVS-----DFAGEFVSGSGTAAYRDAITLASGLVANALEAADAPGLV
MAB_1685|M.abscessus_ATCC_1997 MTAHQSPFVQSPFVSDFLTGDSAHTYREAVALAAERASVTLGGATAPGLV
MSMEG_3506|M.smegmatis_MC2_155 MSRRTAG----------LDVAVSHAKAFLAQLDDRPVAARGDASAVRQLL
TH_2178|M.thermoresistible__bu MSDRTAG----------LTTAAEQAHRFLTGLDDRPVGARADAAAVRGLL
*: : : : * .. .: . *:
MUL_4929|M.ulcerans_Agy99 DGYSLLKELADKPRLDTVAGVGLPAALEEAVGLMAGHSAVVTHPAYLAHL
MMAR_0167|M.marinum_M DGYSLLKELADKPRLDTVAGVGLPAALEEAVGLMAGHSAVVTHPAYLAHL
MAB_1685|M.abscessus_ATCC_1997 DGYSALKRDAERLDLDDPDGVGLSRALDEATGLLAEKSVVVTHPAYLAHL
MSMEG_3506|M.smegmatis_MC2_155 G--APLPETGE-----DPAAVIEGLAVGAEPGLVAG-----AGPRHFGFV
TH_2178|M.thermoresistible__bu D--GPLPEHGE-----DPDRVITALVDGAEPGLVAG-----AGPRHFGFV
. . * . .: * . **:* : * ::..:
MUL_4929|M.ulcerans_Agy99 HCPPTVASLSAEVLISAFNQSLDSFDQAPAATAIEQRVVEHLCARIGYGT
MMAR_0167|M.marinum_M HCPPTVASLSAEVLISAFNQSLDSFDQAPAATAIEQRVVEHLCARIGYGT
MAB_1685|M.abscessus_ATCC_1997 HCPPALPSLAAEVLISAFNQSMDSFDQAPAATAIEQQVVEYLCTRMGYGR
MSMEG_3506|M.smegmatis_MC2_155 IGGSLPAALGADWLVSAWDQCAAFHALSPAATAIEEITAGWVLDLLGLPA
TH_2178|M.thermoresistible__bu IGGVLPAALAADWLVSAWDQCAAFHSLSPAAAAIEEITAAWILDLLGLPD
.:*.*: *:**::*. . :***:***: .. : :*
MUL_4929|M.ulcerans_Agy99 DADGTFTSGGTQSNLHALLMARDVFAQRTLGLDVGTAGLPAAARSWRLLC
MMAR_0167|M.marinum_M DADGTFTSGGTQSNLHALLMARDVFAQRTLGLDVGTAGLPAAARSWRLLC
MAB_1685|M.abscessus_ATCC_1997 ESDGTFTSGGTQSNLQALLLARDLFAR-KAGWDIAGLGLPPESGTWRVLC
MSMEG_3506|M.smegmatis_MC2_155 ECGVGFVTGAQAANTTGLAAARHAVLA-RAGWDVSRDGLIG-APRIRVVC
TH_2178|M.thermoresistible__bu TASVGFVTGGQGANTTCLAAARHAALA-KVGWNVERDGLIG-APPLRVVC
.. *.:*. :* * **. * :: ** : *::*
MUL_4929|M.ulcerans_Agy99 TRQAHFSARQALRILGLGADAVIEVPTDDAGRLCVEALAGCIAESQARGT
MMAR_0167|M.marinum_M TRQAHFSARQALRILGLGADAVIEVPTDDAGRLCVEALAGCIAESQARGT
MAB_1685|M.abscessus_ATCC_1997 TRQTHFSVQQALRLLGLGTSAIIEVPTDDAGRLRAETLAGILRDADHAGT
MSMEG_3506|M.smegmatis_MC2_155 GGQAHTTVYAALRLLGLGEASAARVAVDDQGRMRPEALAETLAAGTGP--
TH_2178|M.thermoresistible__bu GEQAHVTIDTALRLLGLGDSTSVRVPADDQGRMDPQALQRILASSSEP--
*:* : ***:**** : .*..** **: ::* : .
MUL_4929|M.ulcerans_Agy99 PVFGLFLTAGTTDYGAIDPLEPAIKIARQHGVWAHVDACVGGCLAFSAQH
MMAR_0167|M.marinum_M PVFGLFLTAGTTDYGAIDPLEPAIKIARQHGVWAHVDACVGGCLAFSAQH
MAB_1685|M.abscessus_ATCC_1997 PVFALVLTAGTTDFGAIDPLEEATAIARSHGIWVHVDACAGGCLIFSAQH
MSMEG_3506|M.smegmatis_MC2_155 --AIVCAQAGNVATGAFDDLDVIADICSDHDAWLHVDGAFGLWAAAAPST
TH_2178|M.thermoresistible__bu --TIVCAQAGNVSTGAFDDFGPIADACDEHGAWLHIDGAFGLWAAAAPGT
: **.. **:* : . .*. * *:*.. * :.
MUL_4929|M.ulcerans_Agy99 RHLLRGIELADSVAVDFHKLLFQAISCSALLVRDAESFRVSAGHADYLNP
MMAR_0167|M.marinum_M RHLLRGIELADSVAVDFHKLLFQAISCSALLVRDAESFRVSAGHADYLNP
MAB_1685|M.abscessus_ATCC_1997 RQLLRGIEQADSVAIDFHKLLFQAISCSALLVRNRESFGVLAAHADYLNP
MSMEG_3506|M.smegmatis_MC2_155 RHLTAGVARADSWAVDAHKWLNVPYDCAMAIVRDADAHVAAMSLAGPYLI
TH_2178|M.thermoresistible__bu RHLTRGVERADSWAVDAHKWLNVPYDAAMAIVADSEAHLAAMSLAGPYLV
*:* *: *** *:* ** * . ..: :* : ::. . . *.
MUL_4929|M.ulcerans_Agy99 ADDVEHDVVNLVGKSLQTTRRFDALKIFLTLRALGERRIGAMIDATCAAA
MMAR_0167|M.marinum_M ADDVEHDVVNLVGKSLQTTRRFDALKIFLTLRALGERRIGAMIDATCAAA
MAB_1685|M.abscessus_ATCC_1997 ASDAEHDVLNLVGNSLQTTRRFDALKVLVTLRAVGQRQVAAMIDATVAAA
MSMEG_3506|M.smegmatis_MC2_155 VDPGQRDNTNFVPE---SSRRARAVPVYAALRSLGRAGLADLIERNCAQA
TH_2178|M.thermoresistible__bu TDPVHRDSTNYVPE---SSRRSRAVPVYAALRSLGRSGLAELIERNCAQA
.. .:* * * : ::** *: : :**::*. :. :*: . * *
MUL_4929|M.ulcerans_Agy99 AAAGRAVSDHPHLTLCAPVAVNTVVLRWRHPELTEQKCDAVNSAIRSSLA
MMAR_0167|M.marinum_M AAAGRAVSDHPHLTLCAPVAVNTVVLRWRHPELTEQKCDAVNSAIRSSLA
MAB_1685|M.abscessus_ATCC_1997 AAAAQVAAEHPDLELVAPVCTNTVVLRWRRPEATRDARDDVNDAVRAELA
MSMEG_3506|M.smegmatis_MC2_155 RRVARLLAEIPGAQILNDVVLNQVLVRLP-------GGDEATRRAVAAIQ
TH_2178|M.thermoresistible__bu RRMARRLQDIPGAKIRNDVVLNQVLLRLP-------GGDDANRAAVAAVQ
.: : * : * * *::* * .. : :
MUL_4929|M.ulcerans_Agy99 RCGLALVGRSRAAGHQAIKLTFVNPLITADLATGVIADVSRRGNEIAGLP
MMAR_0167|M.marinum_M RCGLALVGRSRAAGHQAIKLTFVNPLITADLATGVIADVSRRGNEIACLP
MAB_1685|M.abscessus_ATCC_1997 HSGRAIVGRSGAAGAQAIKLTFVNPLVTTSMAGAMVSDIAAHGNRIWRAR
MSMEG_3506|M.smegmatis_MC2_155 ADGTCWLGGTTWNDEFVLRLSFTNWATSDDDVDRSAAAITDAVASSRTSQ
TH_2178|M.thermoresistible__bu RDGTCWLGGTTWRGEYVVRLSFSNWSTTDDDIDRSAEAIAVAARSVAAAG
* . :* : . .::*:* * : . ::
MUL_4929|M.ulcerans_Agy99 DDLAAQA
MMAR_0167|M.marinum_M DDLAAQA
MAB_1685|M.abscessus_ATCC_1997 TPEVSAV
MSMEG_3506|M.smegmatis_MC2_155 QPG----
TH_2178|M.thermoresistible__bu -------