For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTAPGDPAASETPLDPSPSADEPRTRIIRRAPTGPQPVTTDSATAKISHRPTSLDETARADRLPPLPDGL PVEPSSRTAVATAAVSIVNGWATSVVATDLITGWWRSGRLFCIAVGFLALVFASSTTAGVIMLLLRRPFG RYLIVVGAVVAVLTYGGVFIAGALVAWIVHAFPVLPIASAVLALHPQTRRWLFD
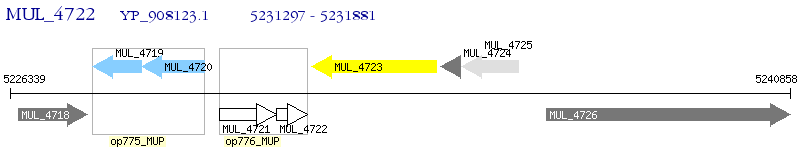
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4722 | - | - | 100% (194) | hypothetical protein MUL_4722 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1863 | - | 6e-40 | 46.00% (200) | hypothetical protein Mflv_1863 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4552 | - | 1e-84 | 98.73% (157) | hypothetical protein MMAR_4552 |
| M. avium 104 | MAV_1072 | - | 9e-61 | 63.96% (197) | hypothetical protein MAV_1072 |
| M. smegmatis MC2 155 | MSMEG_5527 | - | 4e-40 | 45.45% (198) | hypothetical protein MSMEG_5527 |
| M. thermoresistible (build 8) | TH_2201 | - | 3e-32 | 55.14% (107) | conserved hypothetical protein |
| M. vanbaalenii PYR-1 | Mvan_4872 | - | 1e-36 | 46.37% (179) | hypothetical protein Mvan_4872 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5527|M.smegmatis_MC2_155 MSDSGPLDT--------------DQSGEDDRTRIIRRQPSSPLSGPVPTS
TH_2201|M.thermoresistible__bu --------------------------------------------------
Mflv_1863|M.gilvum_PYR-GCK MASDPPSTSSTAPEPGEPAQVPPSDRDEDTSTRIIRR---APLNTGMNTG
Mvan_4872|M.vanbaalenii_PYR-1 MADES------------------SDSTEET--RILRR---APLNTGAAP-
MUL_4722|M.ulcerans_Agy99 MTAPGDPAASETP----LDPSPSADEP---RTRIIRR---APTGP-----
MMAR_4552|M.marinum_M --------------------------------------------------
MAV_1072|M.avium_104 MTAPNDPPSGPRP----SEPAAEPSPPSDTRTRIIRR---APTGP-----
MSMEG_5527|M.smegmatis_MC2_155 DPLSDDVPKTSIIRRHPTGAIPTAPDAEPQTSLIGAQSDASGPEDAGEAA
TH_2201|M.thermoresistible__bu --------------------------------------------------
Mflv_1863|M.gilvum_PYR-GCK SRAAAAEPQTTILRRHPTGALPAL----------------SEPVTQAVRH
Mvan_4872|M.vanbaalenii_PYR-1 -SAPTSEPVTQIVPR-------------------------AQPVAVGP--
MUL_4722|M.ulcerans_Agy99 QPVTTDSATAKISHRPT-----------------------SLDETARADR
MMAR_4552|M.marinum_M --MTTDSATAKISHRPT-----------------------SLDETARADR
MAV_1072|M.avium_104 IPVTPDSPTTHFAPQPP-----------------------SYRGGPRAGR
MSMEG_5527|M.smegmatis_MC2_155 TAYVPRARPLVIPRRASANDPKTAVTAATLSILSGWGTAVIATDLIAGWW
TH_2201|M.thermoresistible__bu ----------------------------VVTILSGWATAVIATDLITGWW
Mflv_1863|M.gilvum_PYR-GCK PEPVPGPPP---------ADPTSAIATTVASIVAGWATGVIATDLITGWW
Mvan_4872|M.vanbaalenii_PYR-1 ----PRAAR---------ADPKSAIATAVTSILSGWATTVIATDLITGWW
MUL_4722|M.ulcerans_Agy99 LPPLPDGLP-------VEPSSRTAVATAAVSIVNGWATSVVATDLITGWW
MMAR_4552|M.marinum_M LPPLPDGLP-------VEPSSRTAVATAAVSIVNGWATSVVATDLITGWW
MAV_1072|M.avium_104 TG--YSGLPA------LEPSDRTAVAACAVAIVSGWATSVVATDLIAGWW
. :*: **.* *:*****:***
MSMEG_5527|M.smegmatis_MC2_155 RTDQLFCVAVGFLTAVSAAATIGGLIALLLRRRLGRLLIVVGAVIALLIF
TH_2201|M.thermoresistible__bu RTDRLFCVAVGFLTAVSAAAMIGGLILMLLRRRLGRLLTVVGGVIGLLIF
Mflv_1863|M.gilvum_PYR-GCK ATDPLFCVAMGFLAAVSAAASISGVIALLLRRRTARLLIVVGAVIALLIF
Mvan_4872|M.vanbaalenii_PYR-1 ATDPLFCVAMGFLAAVSAAASISGLIALLLRRRTARLLVVVGAVIALLIF
MUL_4722|M.ulcerans_Agy99 RSGRLFCIAVGFLALVFASSTTAGVIMLLLRRPFGRYLIVVGAVVAVLTY
MMAR_4552|M.marinum_M RSDRLFCIAVGFLALVFASSTTAGVIMLLLRRPFGRYLIVVGAVVAVLTY
MAV_1072|M.avium_104 RSDRLFCIAVGFLALVFAATTVSGVIGLLQRRSVSRYLLLAGALVAFLTY
:. ***:*:***: * *:: .*:* :* ** .* * :.*.::..* :
MSMEG_5527|M.smegmatis_MC2_155 GSLFIAGAKLHPLVYAIPVLPVSAILFAVLPATGRWAVRSE
TH_2201|M.thermoresistible__bu GSLFVAGAALPPVVYAIPILPVTSIVLTVLPATGRWADLS-
Mflv_1863|M.gilvum_PYR-GCK AGLFVAGAALPGPVYALPLLPAAAIALAALPGTARWSRAD-
Mvan_4872|M.vanbaalenii_PYR-1 ASLFVAGANLPALVYAMPLLPAASIVLAALPATARWSRSA-
MUL_4722|M.ulcerans_Agy99 GGVFIAGALVAWIVHAFPVLPIASAVLALHPQTRRWLFD--
MMAR_4552|M.marinum_M GGVFIAGARVAWIVHAFPVLPIASAVLALHPQTRRWLFD--
MAV_1072|M.avium_104 IGVFIAGARVAWVVHLLPVLPIATVVLVLLPQTKRWLDA--
.:*:*** : *: :*:** :: :. * * **