For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNALFTTAMALREVEAPAGGQCRIFDGELDENWTIGPKVHGGVMLALCAKAARGAHGGDALQPVAVSTNF LWAPDPGPMRLVTSIRKRGRRISVVDVELMQGKRTAVHAVVTLGEPEHYESGPAAPLLSANPVLEMMAPE PPDDVTPIGPGHALAGLAHLGAGCDVRPLASTLQTPASGWGGRAPTFQMWARPRGVAPDALFALMCGDLS VPVTFAVGRTGWAPTVQLTAILRALPTDGWLRIVATCVEIGQGWFDEDHIVVDQAGRIVVQTRQLAMVPA R
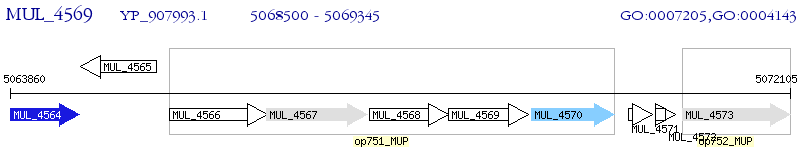
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4569 | - | - | 100% (281) | hypothetical protein MUL_4569 |
| M. ulcerans Agy99 | MUL_4450 | - | 2e-55 | 43.68% (261) | hypothetical protein MUL_4450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0510 | - | 1e-120 | 73.13% (294) | hypothetical protein Mb0510 |
| M. gilvum PYR-GCK | Mflv_0076 | - | 7e-97 | 60.00% (280) | hypothetical protein Mflv_0076 |
| M. tuberculosis H37Rv | Rv0499 | - | 1e-120 | 73.13% (294) | hypothetical protein Rv0499 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4010c | - | 7e-76 | 52.07% (290) | hypothetical protein MAB_4010c |
| M. marinum M | MMAR_0825 | - | 1e-163 | 98.58% (281) | hypothetical protein MMAR_0825 |
| M. avium 104 | MAV_4653 | - | 1e-117 | 73.87% (287) | hypothetical protein MAV_4653 |
| M. smegmatis MC2 155 | MSMEG_0942 | - | 1e-102 | 66.91% (278) | hypothetical protein MSMEG_0942 |
| M. thermoresistible (build 8) | TH_0956 | - | 2e-39 | 74.47% (94) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_0836 | - | 1e-93 | 57.79% (289) | hypothetical protein Mvan_0836 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4569|M.ulcerans_Agy99 ---MNALFTTAMALREVEAPAGG-QCRIFDGELDENWTIGPKVHGGVMLA
MMAR_0825|M.marinum_M ---MNALFTTAMALREVEAPAGG-QCRIFDGELDENWTIGPKVHGGVMLA
MAV_4653|M.avium_104 ---MNALFTTAMTLREAGS--G-----VYDGELDKRWTIGPKVHGGAMLA
Mb0510|M.bovis_AF2122/97 ---MNALFTTAMALRPLDSDPGNPACRVFEGELNEHWTIGPKVHGGAMVA
Rv0499|M.tuberculosis_H37Rv ---MNALFTTAMALRPLDSDPGNPACRVFEGELNEHWTIGPKVHGGAMVA
Mflv_0076|M.gilvum_PYR-GCK -MTSSTLFTDAMALTPAGDG-------VYHGVLNEHWTIGPKVHGGAMLA
Mvan_0836|M.vanbaalenii_PYR-1 MMAGSTLFTDAMALTPAGDG-------VYHGVLNEHWTIGPKVHGGAMLA
MSMEG_0942|M.smegmatis_MC2_155 -MSASTLFTDAMALTPAGDG-------IYDGALDEHWTIGPKVHGGAMLA
TH_0956|M.thermoresistible__bu --------------------------------------------------
MAB_4010c|M.abscessus_ATCC_199 --MVDAPFAQAMSLTPAGEG-------LFDAHLNDTWTIGPKLHGGVMLA
MUL_4569|M.ulcerans_Agy99 LCAKAARGAHGG---------DALQPVAVSTNFLWAPDPGPMRLVTSIRK
MMAR_0825|M.marinum_M LCAKAARAAHGG---------DALQPVAVSTNFLWAPDPGPMRLVTSIRK
MAV_4653|M.avium_104 LCANAARTACAGG------PESAQQPVAVSASFLWAPDPGPMRLVTSIRK
Mb0510|M.bovis_AF2122/97 LCANAARTAYGAAG-----QQPMRQPVAVSASFLWAPDPGTMRLVTSIRK
Rv0499|M.tuberculosis_H37Rv LCANAARTAYGAAG-----QQPMRQPVAVSASFLWAPDPGTMRLVTSIRK
Mflv_0076|M.gilvum_PYR-GCK LCANAARSEFGE---------DGVEPIAVSGSFLWAPDPGAMQVVTTVRK
Mvan_0836|M.vanbaalenii_PYR-1 LCANAARTEFDE---------PGVEPIAVSGSFLWAPDPGPMEVVTTVRK
MSMEG_0942|M.smegmatis_MC2_155 LCANAAQTQIG----------GELQPIAVSGNFLWAPDPGPMRLVTTVCK
TH_0956|M.thermoresistible__bu --------------------------------------------------
MAB_4010c|M.abscessus_ATCC_199 LLAGAARESLTEGLRTSGSSAGQREPIAVSASYLFAPSPGDMQVRTSVRK
MUL_4569|M.ulcerans_Agy99 RGRRISVVDVELMQGK-----------RTAVHAVVTLGEPEHYESG----
MMAR_0825|M.marinum_M RGRRISVVDVELMQGE-----------RTAVHAVVTLGEPEHYESG----
MAV_4653|M.avium_104 RGRRISVVDVELTQGE-----------RTAVHAVVNLGEPEHFPADGS--
Mb0510|M.bovis_AF2122/97 RGRRISVADVELTQGG-----------RTAVHAVVTLGEPEHFLPGVDGS
Rv0499|M.tuberculosis_H37Rv RGRRISVADVELTQGG-----------RTAVHAVVTLGEPEHFLPGVDGS
Mflv_0076|M.gilvum_PYR-GCK RGRRVSLLDVELQQGD-----------RVALRAAITMGTPEH--------
Mvan_0836|M.vanbaalenii_PYR-1 RGRRVSLIDVELRQGVRTRDERSRVEQRVALRAAITMGTPEH--------
MSMEG_0942|M.smegmatis_MC2_155 RGRRISVVDVELNQGA-----------RTAVRASVTLGEPEH--------
TH_0956|M.thermoresistible__bu --------------------------------------------------
MAB_4010c|M.abscessus_ATCC_199 HGKQISLVDAELIQGD-----------RTAVHAVVTLGTPEQ--------
MUL_4569|M.ulcerans_Agy99 ----PAAPLLSAN-PVLEMMAPEPPDDVTPIGPGHALAGLAHLGAGCDVR
MMAR_0825|M.marinum_M ----PATPLLSAN-PVLEMMAPEPPDDVTPIGPSHALAGLAHLGAGCDVR
MAV_4653|M.avium_104 --AASAKPLLSAN-PVPALMPPEPPDDIEPIGPGHPLAGLVHLGEGCDVR
Mb0510|M.bovis_AF2122/97 GGASGTAPLLSAN-PVVELMAPEPPEGVVPIGPGHQLAGLVHLGEGCDVR
Rv0499|M.tuberculosis_H37Rv GGASGTAPLLSAN-PVVELMAPEPPEGVVPIGPGHQLAGLVHLGEGCDVR
Mflv_0076|M.gilvum_PYR-GCK ----HVPPLLSVN-PVVPLMTPEPPPGLEPIGPGHSMADIVHLAHGCDIR
Mvan_0836|M.vanbaalenii_PYR-1 ----HVPPLLSVN-PVIPLMTPEPPPGLEPIGPGHSMADIVHLAHGCDIR
MSMEG_0942|M.smegmatis_MC2_155 ----HVAPLLSFN-PVSPLMAPEPPPEIAPIGPGHPMEHIVHLAHGCDIR
TH_0956|M.thermoresistible__bu --------------------------------------------------
MAB_4010c|M.abscessus_ATCC_199 ----HVGPLLTALPPALTQLPAEPPPGVAPIAEGHPMGEIFHLFGGLDVR
MUL_4569|M.ulcerans_Agy99 PLASTLQTPASGWGGRAPTFQMWARPRGVAPDALFALMCGDLSVPVTFAV
MMAR_0825|M.marinum_M PLASTLQTPASGWGGRAPTFQMWARPRGVAPDALFALMCGDLSVPVTFAV
MAV_4653|M.avium_104 PVLSTMQPSAD---GRPPVIQMWARPRGVAPDALFALMCGDLSAPVTFAV
Mb0510|M.bovis_AF2122/97 PVLSTLRSATD---GRPPVIQLWARPRGVAPDALFALLCGDLSAPVTFAV
Rv0499|M.tuberculosis_H37Rv PVLSTLRSATD---GRPPVIQLWARPRGVAPDALFALLCGDLSAPVTFAV
Mflv_0076|M.gilvum_PYR-GCK PSLTTFDPRQD---GGMPVIEYWVRPKGVAPDLLFALLCGDVSAPVTFGV
Mvan_0836|M.vanbaalenii_PYR-1 PSLTSFSRRAD---GGMPVIEYWVRPKGVAPDVLFALLCGDVSAPVTFGV
MSMEG_0942|M.smegmatis_MC2_155 PALHTLGAREG---GGPPVFEYWVRPKGVAPDVLFALLCGDVSAPVTFAV
TH_0956|M.thermoresistible__bu ------------------VVEMWVRPRDAAPDVLFALMCGDVSAPVTYAV
MAB_4010c|M.abscessus_ATCC_199 PSLRSMSDLSE---RGKPEIVLWARPRDMAPDGLFALVCGDISVPVTFPL
. *.**:. *** ****:***:*.***: :
MUL_4569|M.ulcerans_Agy99 GRTGWAPTVQLTAILRALPTDGWLRIVATCVEIGQGWFDEDHIVVDQAGR
MMAR_0825|M.marinum_M GRTGWAPTVQLTAILRALPTDGWLRIVATCVEIGQGWFDEDHIVVDQAGR
MAV_4653|M.avium_104 DRTGWAPTIQLTAFLRGLPADGWLRIIATCTEIGHDWFDEDHTVVDSLGR
Mb0510|M.bovis_AF2122/97 DRTGWAPTVALTAYLRALPADGWLRVLCTCVEIGQDWFDEDHIVVDRLGR
Rv0499|M.tuberculosis_H37Rv DRTGWAPTVALTAYLRALPADGWLRVLCTCVEIGQDWFDEDHIVVDRLGR
Mflv_0076|M.gilvum_PYR-GCK NRFGWAPTVQLTAYLRALPADGWLRVMCTATQIGEDWFDEDHIVVDCEGR
Mvan_0836|M.vanbaalenii_PYR-1 NRFGWAPTVQLTAFLRALPADGWLRVMCTATQIGQDWFDEDHIVVDCEGR
MSMEG_0942|M.smegmatis_MC2_155 ERTGWAPTVQLTAYLRGLPADGWLRVMCTTVQIGEDWFDEDHTVVDCEGR
TH_0956|M.thermoresistible__bu QRFGWAPTVQLTAYLRGLPADGWLRVICTTMQIGQDWFDEDHVVVDHAGR
MAB_4010c|M.abscessus_ATCC_199 GRFGWAPTVQLTTYLRTLPADGWLRVLCSTNQIGQHWFDSEHTVIDSEGA
* *****: **: ** **:*****::.: :**. ***.:* *:* *
MUL_4569|M.ulcerans_Agy99 IVVQTRQLAMVPAR--
MMAR_0825|M.marinum_M IVVQTRQLAMVPAR--
MAV_4653|M.avium_104 LVVQARQLALVPAAP-
Mb0510|M.bovis_AF2122/97 IVVQTRQLAMVPAQ--
Rv0499|M.tuberculosis_H37Rv IVVQTRQLAMVPAQ--
Mflv_0076|M.gilvum_PYR-GCK IIVQTRQLAMVPSR--
Mvan_0836|M.vanbaalenii_PYR-1 IIVQTRQLALVPTG--
MSMEG_0942|M.smegmatis_MC2_155 IIVQSRQLAMVPTAQG
TH_0956|M.thermoresistible__bu IIVQTRQLAMVPQR--
MAB_4010c|M.abscessus_ATCC_199 LVVQSRQLAMVPVN--
::**:****:**