For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLSEAPKEIAGHGLLADKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERRLGETAAELSALGLGRV ESVLCDVSSTAQVDALIDSTTARMGRLDVLVNNAGLGGQTPVADMIDDEWDRVLDVTLTSVFRATRAALR YFREAPHGGVIVNNASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSIARHKFLDKT TSAELLDRLSAGEAFGRAAEPWEVAATIAFLASEYSSYLTGEVISVSSQHP*
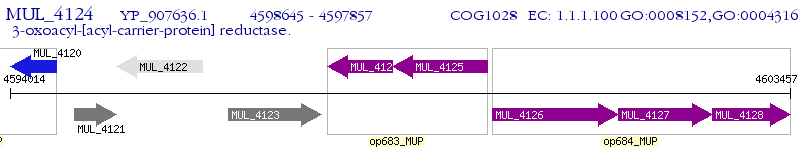
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4124 | - | - | 100% (262) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3898 | fabG | 2e-36 | 38.66% (238) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. ulcerans Agy99 | MUL_0327 | - | 1e-31 | 35.39% (243) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3589c | - | 1e-137 | 94.66% (262) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1482 | - | 1e-122 | 85.27% (258) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3559c | - | 1e-137 | 94.66% (262) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 1e-23 | 35.57% (253) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_0598 | - | 1e-119 | 81.92% (260) | short chain dehydrogenase |
| M. marinum M | MMAR_5048 | - | 1e-143 | 98.85% (262) | short chain dehydrogenase |
| M. avium 104 | MAV_0602 | - | 1e-133 | 91.60% (262) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6011 | - | 1e-120 | 83.46% (260) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2069 | - | 1e-123 | 85.77% (260) | PROBABLE OXIDOREDUCTASE |
| M. vanbaalenii PYR-1 | Mvan_5284 | - | 1e-122 | 84.62% (260) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4124|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5048|M.marinum_M --------------------------------------------------
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
TH_2069|M.thermoresistible__bu --------------------------------------------------
Mflv_1482|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5284|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6011|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
MUL_4124|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5048|M.marinum_M --------------------------------------------------
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
TH_2069|M.thermoresistible__bu --------------------------------------------------
Mflv_1482|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5284|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6011|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
MUL_4124|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5048|M.marinum_M --------------------------------------------------
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0602|M.avium_104 --------------------------------------------------
TH_2069|M.thermoresistible__bu --------------------------------------------------
Mflv_1482|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5284|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6011|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
MUL_4124|M.ulcerans_Agy99 ---------------------------------------------MSLSE
MMAR_5048|M.marinum_M ---------------------------------------------MSLSE
Mb3589c|M.bovis_AF2122/97 ---------------------------------------------MNLSV
Rv3559c|M.tuberculosis_H37Rv ---------------------------------------------MNLSV
MAV_0602|M.avium_104 ---------------------------------------------MSLSE
TH_2069|M.thermoresistible__bu -------------------------------------------VTTDLSQ
Mflv_1482|M.gilvum_PYR-GCK --------------------------------------------MTDLSQ
Mvan_5284|M.vanbaalenii_PYR-1 --------------------------------------------MTDLSV
MSMEG_6011|M.smegmatis_MC2_155 ---------------------------------------------MNLSE
MAB_0598|M.abscessus_ATCC_1997 ---------------------------------------------MNLAE
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
. :
MUL_4124|M.ulcerans_Agy99 APKEIAGHGLLADKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
MMAR_5048|M.marinum_M APKEIAGHGLLVDKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
Mb3589c|M.bovis_AF2122/97 APKEIAGHGLLDGKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
Rv3559c|M.tuberculosis_H37Rv APKEIAGHGLLDGKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
MAV_0602|M.avium_104 APKEIAGHGLLEGKVVIVTAAAGTGIGSATAKRALAEGADVVISDHHERR
TH_2069|M.thermoresistible__bu APKEIDGHGLLTGKVVVVTAAAGTGIGSAVARRALLEGADVLISDHHERR
Mflv_1482|M.gilvum_PYR-GCK APAEIDGHGLLKGKVVLVTAAAGTGIGSTTARRALLEGADVVVSDYHERR
Mvan_5284|M.vanbaalenii_PYR-1 APTEIEGHGLLKGKVVLVTAAAGTGIGSTTARRALLEGADVVVSDYHERR
MSMEG_6011|M.smegmatis_MC2_155 APKEIDGHGLLKGKVVLVTAAAGTGIGSTTARRALLEGADVVISDYHERR
MAB_0598|M.abscessus_ATCC_1997 APTEIEGHGLLKGKAVVVTAAAGTGIGSSTARRALVEGADVVVSDYHERR
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAAR-GIGAAIAEVVARDGARVVAIDVES--
:. . * .*..:**.** ***:: *. . :** *: * ..
MUL_4124|M.ulcerans_Agy99 LGETAAELSALGLGRVESVLCDVSSTAQVDALIDSTTARMG-RLDVLVNN
MMAR_5048|M.marinum_M LGETAAELSALGLGRVESVLCDVSSTAQVDALIDSTTARMG-RLDVLVNN
Mb3589c|M.bovis_AF2122/97 LGETAAELSALGLGRVEHVVCDVTSTAQVDALIDSTTARMG-RLDVLVNN
Rv3559c|M.tuberculosis_H37Rv LGETAAELSALGLGRVEHVVCDVTSTAQVDALIDSTTARMG-RLDVLVNN
MAV_0602|M.avium_104 LGETADQLAALGQGRVESVLCDVTSTAQVDALFASAHARMG-RIDVLVNN
TH_2069|M.thermoresistible__bu LGETREQLAALGLGRVHSAVCDVTSTEQVDALIATAAEQMG-RLDVLVNN
Mflv_1482|M.gilvum_PYR-GCK LGETRDELAALGLGKVDAVVCDVTSTEAVDALITSTVEKAG-RLDVLVNN
Mvan_5284|M.vanbaalenii_PYR-1 LTETRDELAAFGLGKVDSVVCDVTSTEAVDALITSTVEKAG-RLDVLVNN
MSMEG_6011|M.smegmatis_MC2_155 LGETRDQLADLGLGRVEAVVCDVTSTEAVDALITQTVEKAG-RLDVLVNN
MAB_0598|M.abscessus_ATCC_1997 LTETRDELAALGLGKVAAVVCDVTSTEAVDALITESVAALG-RIDVLVNN
MLBr_02565|M.leprae_Br4923 AAEALDETANRVGG--TTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
*: : : * **:: ** : * : *:****
MUL_4124|M.ulcerans_Agy99 AGLGGQTPVADMIDDEWDRVLDVTLTSVFRATRAALRYFREAPHGGVIVN
MMAR_5048|M.marinum_M AGLGGQTPVADMTDDEWDRVLDVTLTSVFRATRAALRYFREAPHGGVIVN
Mb3589c|M.bovis_AF2122/97 AGLGGQTPVADMTDDEWDRVLDVSLTSVFRATRAALRYFRDAPHGGVIVN
Rv3559c|M.tuberculosis_H37Rv AGLGGQTPVADMTDDEWDRVLDVSLTSVFRATRAALRYFRDAPHGGVIVN
MAV_0602|M.avium_104 AGLGGQTPVVDMTDDEWDRVLDVTLTSVFRATRAALRYFRDAPHGGVIVN
TH_2069|M.thermoresistible__bu AGLGGSTPVIDMTDEEWDRVLDVTLTSVMRATRAALRYFRDAGHGGVIVN
Mflv_1482|M.gilvum_PYR-GCK AGLGGQTPVIDMTDDEWDRVLNVTLTSVMRATRAALRYFRDAPHGGVIVN
Mvan_5284|M.vanbaalenii_PYR-1 AGLGGQTPVIDMTDDEWDRVLNVTLTSVMRATRAALRYFRDAPHGGVIVN
MSMEG_6011|M.smegmatis_MC2_155 AGLGGQTPVVDMTDEEWDRVLNVTLTSVMRATRAALRYFRGVDHGGVIVN
MAB_0598|M.abscessus_ATCC_1997 AGLGGETPVADMTDDQWDRVLDVTLTSTFRATRAALRYFREAGHGGVIVN
MLBr_02565|M.leprae_Br4923 AGITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNG-SISKGGRVIV
**: .. : :* * .** **:*.* : * *.. : :** ::
MUL_4124|M.ulcerans_Agy99 NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSIAR
MMAR_5048|M.marinum_M NASVLGWRAQHSQSHYAAAKAGVMALTRCSAMEAAEYGVRINAVSPSIAR
Mb3589c|M.bovis_AF2122/97 NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSIAR
Rv3559c|M.tuberculosis_H37Rv NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSIAR
MAV_0602|M.avium_104 NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYDVRINAVSPSIAR
TH_2069|M.thermoresistible__bu NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAVEYGVRINAVSPSIAR
Mflv_1482|M.gilvum_PYR-GCK NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAVDYGVRINAVSPSIAR
Mvan_5284|M.vanbaalenii_PYR-1 NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAVEYGVRVNAVSPSIAR
MSMEG_6011|M.smegmatis_MC2_155 NASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAVEFGVRINAVSPSIAR
MAB_0598|M.abscessus_ATCC_1997 NASVLGWRAQYGQSHYAAAKAGVMALTRCSAIEAVEHGVRVNAVAPSIAR
MLBr_02565|M.leprae_Br4923 LSSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIE
:*: * .: .*::**:*****:.**:. * .: .: :***:*.: .
MUL_4124|M.ulcerans_Agy99 HKFLDKTTSAELLDRLSAGEAFGRAAEPWEVAATIAFLASEYSSYLTGEV
MMAR_5048|M.marinum_M HKFLDKTTSAELLDRLSAGEAFGRAAEPWEVAATIAFLASEYSSYLTGEV
Mb3589c|M.bovis_AF2122/97 HKFLDKTASAELLDRLVAGEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
Rv3559c|M.tuberculosis_H37Rv HKFLDKTASAELLDRLAAGEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
MAV_0602|M.avium_104 HKFLEKTSSADLLDRLSAGEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
TH_2069|M.thermoresistible__bu HKFLEKTSSADLLDRLADQEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
Mflv_1482|M.gilvum_PYR-GCK HKFLEKTSSSELLDRLAEGEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
Mvan_5284|M.vanbaalenii_PYR-1 HKFLEKTSSSELLDRLAEGEAFGRAAEPWEVASTIAFLASDYSSYLTGEV
MSMEG_6011|M.smegmatis_MC2_155 HKFLEKTSSSELLDRLASDEAFGRAAEPWEVAATIAFLASDYSSYMTGEV
MAB_0598|M.abscessus_ATCC_1997 HKFLDKVTSSDLLDKLSSDEAFGRAAEPWEVAATIAFLASDYSSYLTGEV
MLBr_02565|M.leprae_Br4923 TQMTAAIPLATREVGRRLN-SLLQGGLPVDVAETIAYFAIPASNAVTGNV
:: . : :: :.. * :** ***::* *. :**:*
MUL_4124|M.ulcerans_Agy99 ISVSSQHP---
MMAR_5048|M.marinum_M ISVSSQHP---
Mb3589c|M.bovis_AF2122/97 ISVSCQHP---
Rv3559c|M.tuberculosis_H37Rv ISVSCQHP---
MAV_0602|M.avium_104 ISVSSQHP---
TH_2069|M.thermoresistible__bu ISVSSQHP---
Mflv_1482|M.gilvum_PYR-GCK ISVSSQRA---
Mvan_5284|M.vanbaalenii_PYR-1 ISVSSQHP---
MSMEG_6011|M.smegmatis_MC2_155 VSVSSQRA---
MAB_0598|M.abscessus_ATCC_1997 ISVSSQRA---
MLBr_02565|M.leprae_Br4923 IRVCGQAMLGA
: *. *